lncRNA Structure Prediction
Species Name : Withania somnifera Sequence Id : GBHJ01002324.1
I. Result for minimum free energy prediction
The optimal secondary structure in dot-bracket notation with a minimum free energy of (-753.00) is given below.
1. Sequence-
CACAUUUUUAUGUUAAAAAUUGAAUAUUUGUGGCCUGGACUGCUCUUAUCUUUGGCGUGC
AUCACUUGUUUUAGCCUGUUUAUUAGUGUGUAAUCAUAACUGUCUUACUGAAAUCAGUUU
GUUCUCCUCUACAGCUUGUCAUGGACAUUGGUGAUAAUGUUAGGUCCUUUGGUUUAAGUU
GUAUAGUUAUUUCGUUAAGUUUUAUGAAUAUAGGGAAAAAGCUCAGCCUGAAAUCACUUG
AAAAUUGUGAAUCUGAUUGUUCUCGUGGGUGCCUGCAUAUAUGGUAGUUUGGAUUAAUUC
AAGAUUACUUAAUAUUUGAUAUUGUCUAGAACUUUGGUUUGCUUUAUUUACUCCAGUAAG
AAUCCUAGGGUCCAAGGAUGCCACUAUGUGAUUUGAAAACUGAGUAUUCAUGGGCUGCUA
GUUUACCUUUUAACCAACAUAAAACUCCACCACUUUAUAUUCAAGCCUAGUGGGAAUGAG
UCCGACUUUUGUUUUCCAGAAACUACACUAAGAAUGGUUUUUUGACUUCAAAAACAUUUG
CAUUCAUACCCCUAACAAAAUGUGAGUCAAAGUUGCAAAAUAUUGAAUAAUAGUUGUUUU
UCUCUUUGUCUUUUUUCUGUAGCUGUUUGUUUUAUCUUGUUUAGGUCGAAUAUAGUAUGG
GCUCAAAUGUUUUAUUUGGUUUAAAAAUGAGUAUAAAACAUGCCUAGGCUAGUGUUCCAG
UCUCUGAAUAUUGUGUUCUUGAUGUUUUAGUUCUGUUUCCCCUUAUUGAAAUCUAUGUGU
ACGAAUGCUAUGUUCCUUCAAAUAAUCAGUUGUAUUCUAGCUUAGCACUUGAUUGUACUG
UAAGUCGGAGCUGUUUCAAGUUGCAAUCUUGAGUGUGUUUAGGAUUAAAAGAGCUAAAAG
CUGAAUCUUCAGGUCACGUGUAGUUCAAUGUCUCCCUUGUUAAUCUCCAUGUUUUAGUUU
UAGAAGUCUAUUUGUCUCAACUUGUUUUGUAGGGUUGCCCAUCAUCCCGAUUUAUGGUCC
UCUAAAUAUGUUUAGUUAGAGUUUAAUUCUAGUAUCUUACCUCUUAAGGCUAUCAUGAAU
AUAGUUUGAUCCCAUGAGUUUUCUUGUUUCAUUUUGUGCGCUAUGUGAUAAAAAUAGUAA
ACUCGUAAGGUUUAUUGGUCGGAGUAGCAAAAUUUCUGGAAUUUGGAAGUUUACAGGGAU
CAAUUCUUAAUCAUGAAAAUUUUCUCAAGUAUGUUUGUGUUCCAAUUUGUUUUCGUUUAG
AAGAAGGAUUGUUAUCACUAGUGCAGUAUAUGUUUUGAACCUCUCUUGUUUUAAGUGCAU
GACAUAGGGUAUGAAAUGAGUAGGAAUGAGCUGAUGGGUUAUUUGUCUUAUGACUAGAUA
UAAGUGUUUCCCUGUACAAGUUAGUUUAGAUAUUGCUUGAAUUGACUGAUUCUUUUGGUG
AACUGUAUGCUUGAAAACUGUGCAAAUAUGCAUUUGUCUGGUGUCUCACUGAGUAGCAAU
CUACCAAGUUUGAGCCUACUUGAGCUAGUUAUAAACAUGGCUAGUUGAUAUAUAAUUGCU
UUCAUAAAGAUGAGGUAGUUCCAUGGGCAUUCGAGUAAAUAGGUCUCUUUUUUAAAAAUA
UAGAUUUAAAUGGGUAAAAUAGGGGUCAUCAAAUGUGUCUGAUAACGCUGGGUGUAUGCA
CUUGGAAUAGACAUAGUUUCCCUUCACCUGGUAGUUUAUUUUUAAUCUGUAUUGAGUAUG
UCAGCUAUAAAAUGUUACUAUGCUUGCCCACCUUAGCUUUUCAUUCGUGUAAAUAUGCUG
AAUAAGGUUGAUUUCUCUUUAGGUGACUUUAAGACUUAGCGACAAAAUGUGAACUUUGAU
GAAUUUGAAUAAUCUGGCAUAGGUUUUCUUCCCUAUCUUGUUUACUUAUCCCAUGGGAAA
AGCAUGCUCUAAAAUCUGUCUCUAAAUAGUGGAACCUGAAUUCAUGCUUAAAUCAUUUCC
UGGAUCUUAAUGGUUAAAGCUUGCCUUGCCUGUUGUGAUAUGUGAAAUCUGCCAGAGACA
UCUGCAAGCUUCAUUGAAAUCUUUUAUUUGCUUUGGUACGUCUUAAGUUUUUUUUUAUGC
CUAUAACUUGUCUAGAUGUUUCAAAUUGGUUUGGAAAUGCGGGAAUUUGUUUUAGAAGCU
CAAAUCUGUUUCUAUCUGUCUACUAGGGCCUGUUGUCUUCAAGCCUUAAAUUCAAAUGGG
AUGAACUCUUUAUCUGGUUCACCUCUGUUCAUAAGAUCAGUGUGUCUUGAGUGUUUAAAA
UGUUAUCACUGCUUGUCUCGUUCAAGUCUUGUCUCUCUUAUGCAACCCAUUUACAAUAGA
UGAAUCAGUUGGAACAGGUUACUGCUAAUGAGUAACAUGAAAUGCAUGCUUGGUCAAAUA
AUUAGGUUUGCUUGCAUCUGAAUGAGUUACUAUCCCGUGGAGGCUGUCAAACACUAAUUU
GAUAAUAUGUGAAUCAGGGAAUAUUUAAAUUUUUACAGUGGUAGUAGGCUUAAAGUCCUC
UGUACAGUAGCAUGAUGAGGUGCUCGGGUUACGAUAUUGAUAGCAUGAUGGGCUGCCUAU
UUUCUCUAUGCAUGGUUUGAGUUUUAUUCUGUUGCCUCUAUGCAUAACUUGGUGUAAUUC
GUUCAUCUCUGAUCAAAUAUAUACGUAGAGACAUAGGACUUAUAAGCAUGUCUUAAUUAG
CUUAAUAUGACUUGAUUGAUACGCUUAUCCUUUAACAGUAAUUGCUUAGCUUAAUCAUAA
CUAAGAGUAGUUAUCUUAGAUUAACAUUUCUGUAGGAUCCUUGAAUUAAUGGAUUACAUG
AUGGCUACAGUUUAGUUAAUGAAAUAUUCUGCCUAGUUGAGCUAGUAUGAAAUACUUGAG
CAUGCAUUGAUUUGAUAAGCAUGAUUGCCCUUUUCUAUUGCGCUAUCUUUACUUUUCUCU
GUUCUUUUCAGUAUUUGACACAAUAUGGUGUCGUUAUUUUUUCUUUUUAUUUUUAGUUUA
AGAUCAUGAUUUGAUUUGAGUAUUAAUUCUUUCUUUUUUGAAUGAUCCGCUUUAUUUGAA
UCCACCGGACUCCCCUUCGGUUUGCACUCCUCGACCGGCAAAGCCCAUCUCUAAAGAGUC
AAGCUUCAUCCAAGCCCAAUUCGGUUGGAGACAUGAAGACUAAUUGGCGCCAAGCCGAAU
CAGAGGCCUUAUCCUUGACCUUCUUUGUAUUAUGAUGUGUAUGUUUACUUGUAUUAAUUU
UAAUCUGGAAUGAAAUCCCAGCCAGUGGCUAUUUAUGUUAUUCAUUUACUCCAAAUUUUC
UUACCUAAAUCGAACUUGUCAUUAAUGUCAAAACAAACGAUUCGUUGAUUAAGUGAA
2. MFE structure-
((((((((((...))))))).........(((.((((((((((.((((((((((....(((.....((((((((((((((((((((((((....))).))))....((((....))))........((.((((((((((((.((((....((....)).....))))..)))..))))))))).))....(((((.....
...)))))((((((((((..(((..........((((.........))))(((.((((((..((((((..((((.(((((((((((.((((((((.......((((.((((......((((..((...((((....))))))..))))........))))))))....))))))))..))))).))))))((((((((((
..(((((((.((.((............((((((((((.(((((((....(((..((((((((..(((((..(((.((((((.(((.(((((...((.((((((((.........)))))))).))...)))))....))).)))))).)))..(((((...(((((.((((((((((.((....((..((.((.......
.)).))..))....)).)))))))..))).))))).)))))))))).))))))))..)))........))))))))))).))))))..(((..((((((((.....(((((((((((........)))))))).)))....)))))))).))).(((((........)))))....)).)).)))))))..)))..))))
)))....(((((.(((((((((.((((.........((((((((((((((((......))))((((..(((((........(((((...((((((((((..((......((((..((((........))))..))..))........))..))))))))))...)))))......))))).))))......(((...)))
......))))))))))))..........)))).))))))))).)))))..................))))..))))))..)))))).)))(((((.(((((((((..((((((((((((.((((((.((....((...((((.....(((((((((((((...(((....((((((((.....)))))))))))...)))
)))(((........)))...........((((....(((...((((((.(((((.....))))).))))))....)))..))))...........))))))).....))))...))..)).)))))).)))))))))))).))))..))))).))))).((((((((....))).)))))..))).))))))))))..((
(((((((((.......)))))))))))((((((((((((((.....(((((.((.....(((((....)))))...)).))))).)))))))).)).)))).....((((((((....))))))))....))))))).))))))..))))......))).....)))))))))))))).)))).)).)))(((((.....
((((((((........((..((((..((((.(((.(((((((.((.(((((..((((((.(((...((((((((....)))).((((((((...(((......((((((((.((.......(((((((.((((.(((((((..((.....((((.......(((..........)))....))))...))..))))))).
))))))))))).....))))))))))....((((..(((((.(((...(((((((..((((((..((((((....((.(((((........))))))))))))).(((((((((..((((((((((.((.....((((.((......))))))....))...)))))))............((((.(((((((....(((
((((..((.((...(((((.......))).))...)).))...)))))))))))))).)))).........(((((.((((((((((((...............))))))...)))))).)))))..((((((((...(((((..((.((..(((((((........))))))).......)).))..)))))....)))
))))).....)))..)))))))))....)))))).)))))))...........(((((((((((((((((((((((...........(((((((((.......(((((((((.(((((.(((.....((((((...)))))).(((((..(((....((((((.......))))))(((....(((.(((((((......
))))))).)))...))).((((((((((((...((((((((((((((((((......)))))))..........(((((((.(((((((((.....((.((.(((((((....((((((((((.(((((((((......))))(((.....))))))))...))).)).)))))))))))).)).))......)))))))
)).)).)))))..)))))))))........)).))))))))))))))))))))..........)))))))).)))))))))))))))).))....(((((((((((......(((((((...(((......(((....)))......))))))))))((((((((.(((((((((.((((........))))...(((((
..........)))))....)))))))))..)))))))).................)))))))))))...))))))))))).)))))))))))).....(((....))).........)))))))))))).)))....))))))))...))))))).))))))...))))))).))))))).))).))))..))))..)).
((((((.....))))))((((....)))).......(((((((((......)))))))))...((((((......)))))).((.((((...((..(((...)))..))......))))))((((((((((((.((......)))))))....))))).))..(((((.....)))))...))))))))....)))))..
...(((((((((((((..((.((((((...(((..(((((.......((((((......((((...)))).......)))))).........)))))....))).)))).)).))..)))).))))).))))...((((...))))......)))..
You can download the minimum free energy (MFE) structure in
II. Results for thermodynamic ensemble prediction
1. The free energy of the thermodynamic ensemble is [-817.52] kcal/mol.
2. The frequency of mfe structure in ensemble 3.44499e-46.
3. The ensemble diversity 1021.57.
4. You may look at the dot plot containing the base pair probabilities [below]
III. Graphical output
1. A note on base-pairing probabilities:--
The structure below is colored by base-pairing probabilities. For unpaired regions the color denotes the probability of being unpaired. The reliability information of RNA secondary structure
plot in the form of color annotation, Positional entropy ranging from red (low entropy, well-defined) via green to blue and
violet (high entropy, ill-defined).
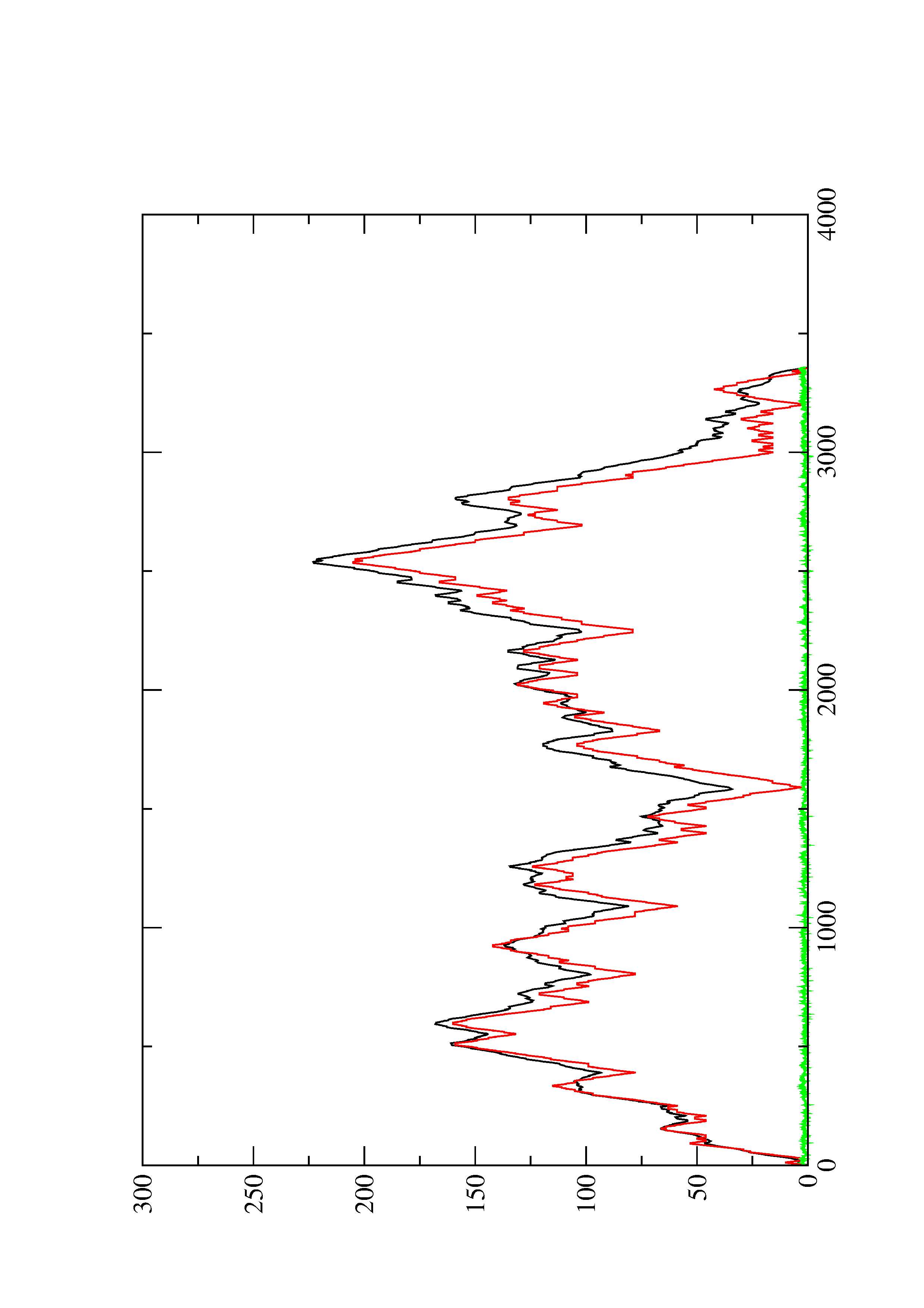
2. Here you find a mountain plot representation of the MFE structure, the thermodynamic ensemble of RNA structures, and the centroid structure. Additionally we present the positional entropy for each position. The resulting plot shows three curves, two mountain plots derived from the MFE structure (red) and the pairing probabilities (black) and a positional entropy curve (green). Well-defined regions are identified by low entropy.