lncRNA Structure Prediction
Species Name : Withania somnifera Sequence Id : GBHJ01002956.1
I. Result for minimum free energy prediction
The optimal secondary structure in dot-bracket notation with a minimum free energy of (-564.80) is given below.
1. Sequence-
GAAGUUUUUAUGUGCAUCUAGUGUUACUUGGGUGUUUUCCUCGUUGAUUUAGCUAUUCUU
UUGAUCUAUAUGUUCUUAUUUCCGAUUUGCUUUAUGAGAAUCUACGGUUAUUUGGGCGUU
UCCCCUAUCUGGGUUUUCAUUGAUUUAGCUAUUAUUUUGGUCUAUGUUGCUCGUACUCUU
UAAAAAUGUCGACUGGUGCGGGUAAGUAGUGUAUUUUUUGAGAAUCCUACAUGUGCGUGG
CAGUAUUUUUGGGGAGUCCGAGCAAAAUAAUUUUUGAUUCAUACAUUCUUUCUGUAUGCC
AUUUUCCCCUUUUUUUGUGUGUGUGUAUGUAUGUUGAAGCUGAUGUUGAGGAGUUCAUUG
CUUUUGGUCUCAGUUCAUGGACCAACUGUAUGCCUUCAUUUCCAUUUUUUCUAAUUUUAU
GUGCAUCUAUAGUUAUCCUUACCUGGCUUAUCAUUGAUUUAGCUAUUGUUUUGAUCCAUA
UGUUUCUGUUUUUAUGUCCUUUUAGCCUUUAGAUUGGUAGAUUCUCUUUAUUGAUUCAAU
ACUCAUUUCUUGGGGUACAUGGCAAUAUCAUUCUCAUGGUCGACAAUUUUCCUGCCAUAC
UAGAAUUCCUAUUAUUUAGAGCAGAGGCUUCCCCAGAAGCAAGCAACAUGAGGUCACUCU
GCUUCGUACCAUAGACAAGGCUCAUUCCGUACUUUACUUAUGAGCUCAUGUAGAUUCAGG
CAUGCUACACUUUUUAGUUAUUGGGCUCAAGUACUCCUUGUCUGUUUGCUAUUGUUUAAA
GUCUUUUCCUUCGGGCGGGUAAGUUUUUGAGAAAAUGGAGUUUUGUAGUCAUUGGUCCUU
UGAAAUCUUUGUUGCUUGGAAACAUUUUAGGAUAGGAACUUAUGGCAGUAAUAUCAUUUG
ACCAGGAGAAUGUGCUUUAGGAUGCUGUUAUGCAUUGAGUGCCUUCAUUACUGUUAAGAU
GCUGUUAUGCUUUGAGUGCCUCCGUUACUGUUAGGAUGCUGUUAUACAUAGAGUCCCUCC
AUUACUGUUAGCUGCUUGUUUGGGUACUUGUUUGAUUCUAGAAAAGUUGGGAUUUUGGUG
UUUAAUUGAACUUUCUUUUAUGUUUGGCUUAAUUGCUUCAAUAACAGAAGUAUUUCUUUU
CAUUCUCCGGAAUUGCGUUUUAUCCAUUUUGUCCAAGUUUGAACUUAAUAUGUAUUUCGU
GAAUCUUUAAAAUGCGUUUUUCUCUAUGAUCAAUUAACUGUAUCAUGAAUGUUCUUAAAU
CGAGUAGAUACGUGAUGAUUAACUUGUUUCUGUUGCAUUCUCAAUUUCUGCCAAAUUUAC
GACGGAAACUUUGCUUCUUCAGUUCCGGUUACCUACUGGUUUAGAAAACUUUGUUUGCCU
CUACCGAACCAUGAUGAUUGACUUUCAUCCUUCAUAUUGCUUUGGAAUCUAAGCUAACCA
UAUGUUUAAACCAUUUAUUGCUUUAUAUCUUUUGGCUCUUGAUUUCUGUGUUUUGUGAUC
UUUAAAGCUGGCUGUUCAACAGCCCGAACUUCUACGGAAUUCUUUUUAAUUAAGGAGAAG
GAGUUUGUUGGGAAUUAAUGUUGGAGUAACUAUGUAAUAUAUGAAUGCACUAUAUAGAAU
AUUAGCACAUUCUGGGGUUGUGCUUCACAUAUCAAUCCCGAGCUUAAUAUAGAAAAUGGU
UGCAGUGCUAAUGGCUAUAGCCUAUAUGCUUUAUUCUCUUUAUUAAAAAGUGGCUUUGUG
CUUUAUUCAUCAUAUCUUUCCUAAUUGGAAAAGGCUCUCAGCUAGAGGGGCAAAAGGAAA
UGGGGAAGGCAACACGGAAGAAGGAUUGAGCGACAUGGAAGCAACCAGAGAUAUUUCCUU
UCUUUUGUGUCAUUCAGAAGAAUGCAGGAAAGAGUGAUGGAAUUCUCUCUUUAAUGGUUG
AAAUAAAUUUGUAGUUAAUGCCCCUUAAUUUUUUCUCUUCAGGCAAGCAAAACUUUCUUU
CCUUUGUCUCACCGCACUUGUUGGGACGUACCAGGACGGGACGGGAUCCCGUUUUCGUCC
UAGUCCGGUCUGAAUUUUUUUUUUGUUAAAAAAAAUAUUUUAUUUAAGAAACGGCCACUU
UUGCAAAGGACAACUAUUCAGGGAUUGGAUUAACGCAGAUCAGCGUAACAAAGGACUCCA
AGGUUAACAUCACAAGAAGAAGCUAUGCUCAAUUUAUUCAUUGGUAACAAUAGUAAUGAU
GGCAUGAAACACGAAGAUCGUCUACAAAAUCUUCUAAUACUAUCUGUUGAUUGCGAUAAG
UCGUUACAAGAUUUAGAUGAGGAAUUUGCUAGACUAUACAACUAUUAAUAUUACAGAACC
CUUC
2. MFE structure-
..(((((((.((((......(((((((((((((.((((...(((((((((.((......((((((((((((((((((.........((((((...(((((.....(((((...((((((...(((.....)))............(((((((...((((.(((((((((((((.(((((((((((((....((((.((((
.(((.((((...(((((...))))).)))).))).)))).)))))))))))))))))..((((((((....))))).))).....((((((((((.((((...((((((((((((((((((((((((((((((((((((((((....((((((((((.(((..((((((.((.....))))))))..))).(((.....(
((((.((((((((((((((((((.....(((((.(((((.((.(((((.(((...)))..)))))........((((..((((((((........................((((((((((.....))))))))))(((((...(((((..((((..((((((.(((..((((.(((..((((.(((((.(((((((((.
(((.....))).)))...(((.((((((((...(((.(((((((...(((((((......))))))).....((((((((........(((((.......(((((((((((((...........)))))............))))))))))))).))))))))))))))).)))....))))))))...))).)))))).
))))).))))....)))))))..))).)))).))..)))).)))))...)))))...))))))))....))))(((.(((((.((((.(((((.(((((...(((..(((.((((((...(((((......))))))))))).)))....)))...))))).))))))))).))))).)))........)).))))))))
))..))))))))))...((((.....((.....)).....))))..............)))))))).)))))....))).......))))).)))))......)))))))..(((((((((((((((((((...............(((((((((...........(((((((.(((((((........((((((((...
.........)))))))).......((((((((......)).))))))................(((((....(((.(((((((........)))).))).)))...))))))))))))..)))))))............)))))))))........(((((((((.......(((.((.....(((((.((..((((..(
(......))..))))..)).))))).......)).)))......)))))))))......(((...........))).....))))))))((((((.......)))))).((((((...)))))).....(((((..(((((((((((........)))))))))))..))))).....))))))))))).....))))))
))..)))))).)))).(((((..((((((....(((((((............)))))))((((((((((.(((.(((((..(((........(((....)))....))).))))).))).)))))......))))))))))).)))))........(((((.....)))))...(((((.....))))))))))))))..
.))))))))))))).)))))))....)))))))))))))..((((((((((...((((.(((((((.((.(((((....))))))).)))))))....))))...)))).....))))))...))))...)))))))))))))..)))))..)))))...))))))((((((...((((((....((((((.((....))
.))))))..(((.(((((((((((((......))))))))).)))))))....(((((((((.....)))))))))...........)))).))....)))))).)))))))..)))....)))))))).....)))))))))))....)))).)).)))))..))))))))))...)))))))(((((....(((((.(
(((....)))))))))....)))))..............(((((((((.(((((...((..((((..((((.((.....)).))))..)))).))...))))).)))).)))))..............................
You can download the minimum free energy (MFE) structure in
II. Results for thermodynamic ensemble prediction
1. The free energy of the thermodynamic ensemble is [-609.96] kcal/mol.
2. The frequency of mfe structure in ensemble 1.51444e-32.
3. The ensemble diversity 753.38.
4. You may look at the dot plot containing the base pair probabilities [below]

III. Graphical output
1. A note on base-pairing probabilities:--
The structure below is colored by base-pairing probabilities. For unpaired regions the color denotes the probability of being unpaired. The reliability information of RNA secondary structure
plot in the form of color annotation, Positional entropy ranging from red (low entropy, well-defined) via green to blue and
violet (high entropy, ill-defined).
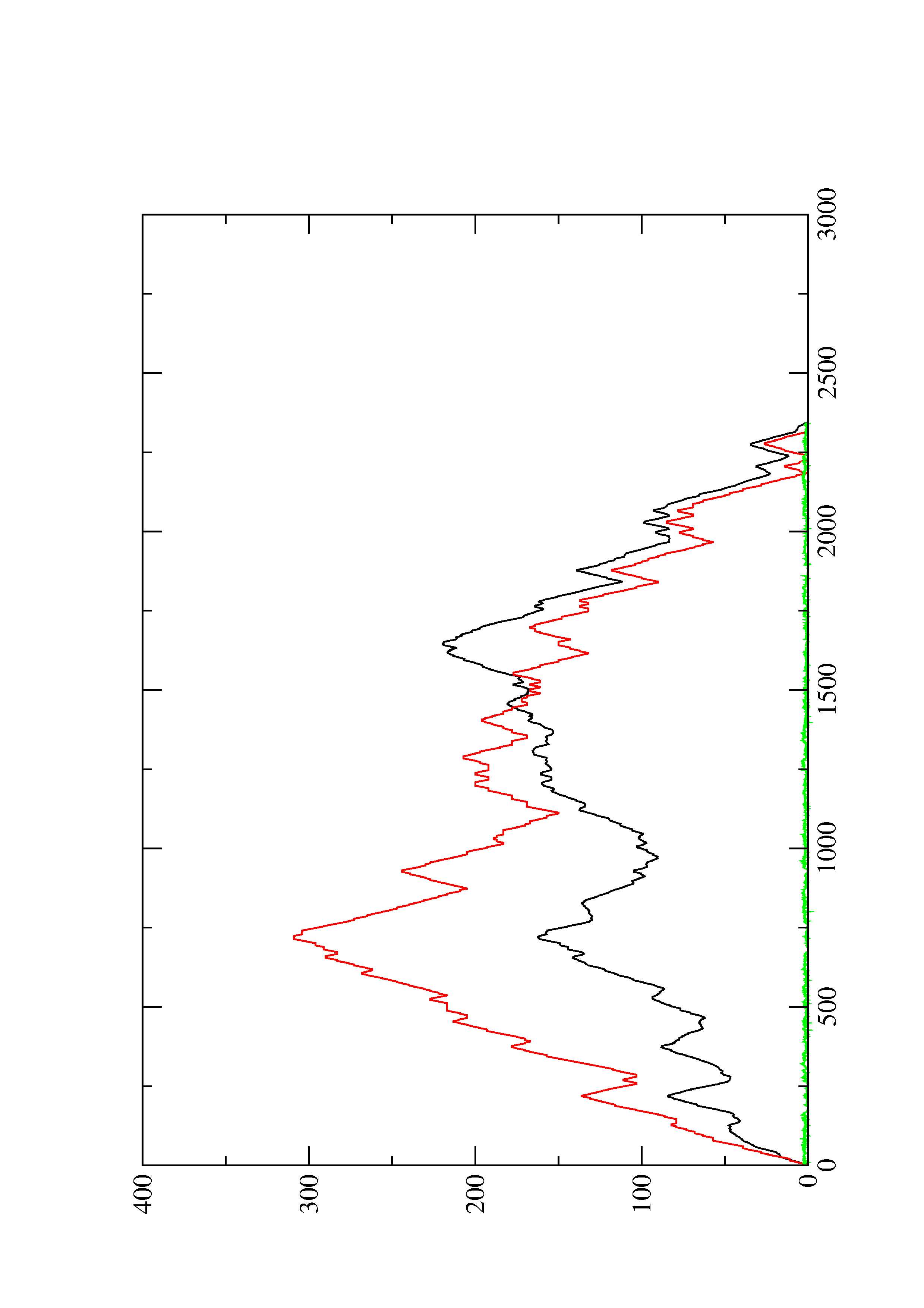
2. Here you find a mountain plot representation of the MFE structure, the thermodynamic ensemble of RNA structures, and the centroid structure. Additionally we present the positional entropy for each position. The resulting plot shows three curves, two mountain plots derived from the MFE structure (red) and the pairing probabilities (black) and a positional entropy curve (green). Well-defined regions are identified by low entropy.