lncRNA Structure Prediction
Species Name : Withania somnifera Sequence Id : GBHJ01003392.1
I. Result for minimum free energy prediction
The optimal secondary structure in dot-bracket notation with a minimum free energy of (-547.30) is given below.
1. Sequence-
AACAAAGAAGAUCAAAAAAGAAAAGUAUUUCGCCUAAGAAUAGGAAUGGAAUGGACCAUC
GAUGGAAAAUAGAAGGUAGACGAGAAGAUAAGGGGCGAAGGAUAUUGGAAAUAGAUUUGG
UCUGUUUCAAAAGGAAUGAAGGAAGGAGUCGACGCAUUGUUUAGCGAUUUAAUGCUUCCU
AUUUUGCCGGGUCCCUCGCCUAGGGGCCAGAAAGGGCAACUUCCUUUCCAGCUUUAGGGC
GGGCCCAGCCUGGGCUUCCGCAAUCGAAGAAAGGAAAGAGCGAGAUGGUGGUUUUUGUUG
UUCCCGCGAAGCGAAGAUCAUUCGCUUGGGACCAAUAUGUGUUAUAUUUACUCACAUAUA
AUGUAAUUUUAACUUACUGAAGAGAUGGCAAGUUGUACUGGCCCUUUUCAGAGGAUUCUU
GCUUCUUUGACCAUUUAUAAUGAUUGCUAUUGGUAGCUCUAAGUAGUAAAUUUGUGUGUU
GCGUUCAAACUUUAGACCAUUUAUGGUCUAAAGUUUAUUCUUCUUGUAGCGUUGAAACAA
GAGAGAACAGUUGAAGGAUGUUACUCAAAUCUCCAAAACUUAUAUUAAUUCUUGAUUUUU
GAAGAAUUUGUUUUCCAAGAGAAAGUGUUUUUCAAAACAUUUUGACGAACCAAACAUGGG
AAAAAUACACCCUCAAUGAAAUGAAAGAGGGUUAAGUCUAUUGACCAGUGUUCAGGAAAG
UGUGAAGCGGAAAAAAGCGACAAGGCCCUGCUUCACGCUUUAAGCAGAGCGUGAAGCGAA
GCGCUCGCUUCAUUGAAAAAGCACACUUUCUUAAAAAAAAUACCAUAAAAUAAAUAGACA
AUAUAUACAAGCAAAAGUUCAAGUUAAAAGCUAAAUAAAGUUCUUCAUAGACCAUAGGUC
AAUCUAAAUCGAAGUUAACGAUGUUUCAGGACAUCUAAUUCUUGUUCUUCUAGUGAUUUA
GUUCAAGAUCGUCAAGAUGUUCUGUUCCUCUAGUAUCAUUAUAUUGCUCAUCAUCUUUCU
CAACUAGUCUUGAAGUAGCUACUUCUUUUUCCUUUCUUAUGUAAGCUUCAAUUAUUCCCC
UUAAAACCACGACGACUUUCCCCAACUCCGCUUGCCUUAGCAAAACACUCCAAAUGAAAU
CAGAGCAGAAAGGAAAACAGAGGAGAAGAGAAGUCCAGAAAACAGGGGAGAAGUGCAGUA
CUAAAGAAAAGGAGCAGAUAAGAAAGAAAGAAAGGAAAGAAGAAGAAGAAGAAAGGUAGA
AGCAGAUGGGUCGAAGCAGAAGACAGACGAAGAAGAGAAGAAAAAAGAAAGAGGAAUAAC
ACUUGAAGAAGAAGUCGCAGCAGCUCCAGUUCCUCAAGUCGCUGCGUUUGUCAACAUAAA
AACCCUAGUCUUGAUUCAAUGUCUUUUGGAAAGGAGCAUACGCCUCAAUUUGCUUCUCGC
CUCAACACAAAAAGCAGUCUCCUUACAAAACCUACAUCACCUCACUACAGUGAGGCGACU
UACCCUCACCUCGCUUCGCCUUGCCUCGCUUCAUUGCUUCCUCGCUUAAAGCGAUGGAGC
GAACACUUUUGUGAACACUGCUAUUGACUGUCCUGCAAAGAGUUGCAACCUGGUUGAACA
CAUCCAUGACUAUGGUAUUCUUUACCAACUUGAUCUUUGAGUAUCAUUCAUUUGUUUUUG
GAGUCUGCAGCUUCUCAAGGUUAUUGAGUGCAUGAAUCAACUUUUCUAGCUCCUUCACUA
GUUUUCAACUCCAUAAUUUUUGUGACUGCAAUUAUUCCCUACUCUGAUAAAAAAGAUAUU
CCCAAUUCUCUGUGCUUCGCUAAGGUGUUCCUGGUUCUUUUUGACAGAUUCAUAAUGAAG
GUGGGCGAGAUCUUCCAUUGGCUUGCACUAUUGUGGACCAGCUUCUACGAGAAGCUGUAG
UCCCUUUGUGAUGUCAUUUUAUGUAAAUGAACUGUUGUAGCUUGUCUUCCUCAACAUCAA
CUUAUUAUGGUCCCUUCUUGAUGAUAUGGUAUCUGUUAUGGUUGUAUUGUUACUUGAUUG
CCUUUGGAUACUUGCUGUGCAAUCUAAAACUUUUAUAUGCUUAUGUUUAUCUCAUUACAA
GUCUGGUCAUGC
2. MFE structure-
..(((((..((((((....((((....)))).((((....))))...((((.(((((((.(((((........(((.(((((((.......(((((((((((((((((((((((((...)))))))))...........((((((.....(((((........))).)).....))))))...(((((.(((((((....
.)))))))......)))))....((((((..((((...(((((((((...)))))..))))....))))...))))))..((((((((.(((((((((((.(((((...((((((.....))))))))))).))(((((((...........))))))).............(((.((((((((..(((.(((...))).
)))))))))))))).((((.(((((...((((...(((((((((.((((..((.(((..(((((.....(((((.(((..(((((((((((((((((((....))))))))))))).((((((((((.........)))))).))))........))))))))).))))).......)))))...............(((
(((((((...(((((...)))))...))))))))))((((((((((((.(((....))).........((((((............))))))...(((((((.....(((((.(((((((((((......((..((((((....((..(((((((((((((....)))))))))))))..))..))))))..))......
.)))))))))))......))))).........)))))))..........(((....))).........((((......)))).......((((...))))...................((((((....))))))....(((((..((((....))...))..)))))))))))))))))...)))))..))))))))))
))).((((..((((((((((....(((.((......)))))(((((((((((((((((((....(((((......((((((.........(((..((((......((((.((..((((.((.......................))...)))).....)).)))).))))..)))........))))))..((((...))
))........)))))..)))))))))............)))))))))).)))))))).))))))....)))))))))))))((((((((.((..((..............(((((((...((((.......)))).((....))....)))))))...)).)).))))))))...)))))))))))).)))))...))))
)))))).....(((((((............))))))))))))).........((((((.((.((((((((...(((((((.........(((((((((............)))))))))(((((((..((((((((((((.(((((...))))).)))))(((((((((.((((((((....((((.......(((((..
..)))))...(((.((....)).)))......(((((.....)))))...........(((((((.((..((((((...(((((.(((((..((.(((((((((.((((...((((.........((((........))))..)))))))).)))).))))).)))))))...))))).......)))))))).))))))
).)))).....))).))))).)))))))))(((...........)))......)))))))..)))))))......((.(((......))).)).((((((((((((...))))))))..))))....)))))))...))).)))))..))))))))....))))))).)))..........)))))))))))).))))((
(((((((...(((......))).))))))))).))))))..)))))((((((((..(((..((...((((..............))))))..)))..)))))...)))....
You can download the minimum free energy (MFE) structure in
II. Results for thermodynamic ensemble prediction
1. The free energy of the thermodynamic ensemble is [-588.16] kcal/mol.
2. The frequency of mfe structure in ensemble 1.6183e-29.
3. The ensemble diversity 470.06.
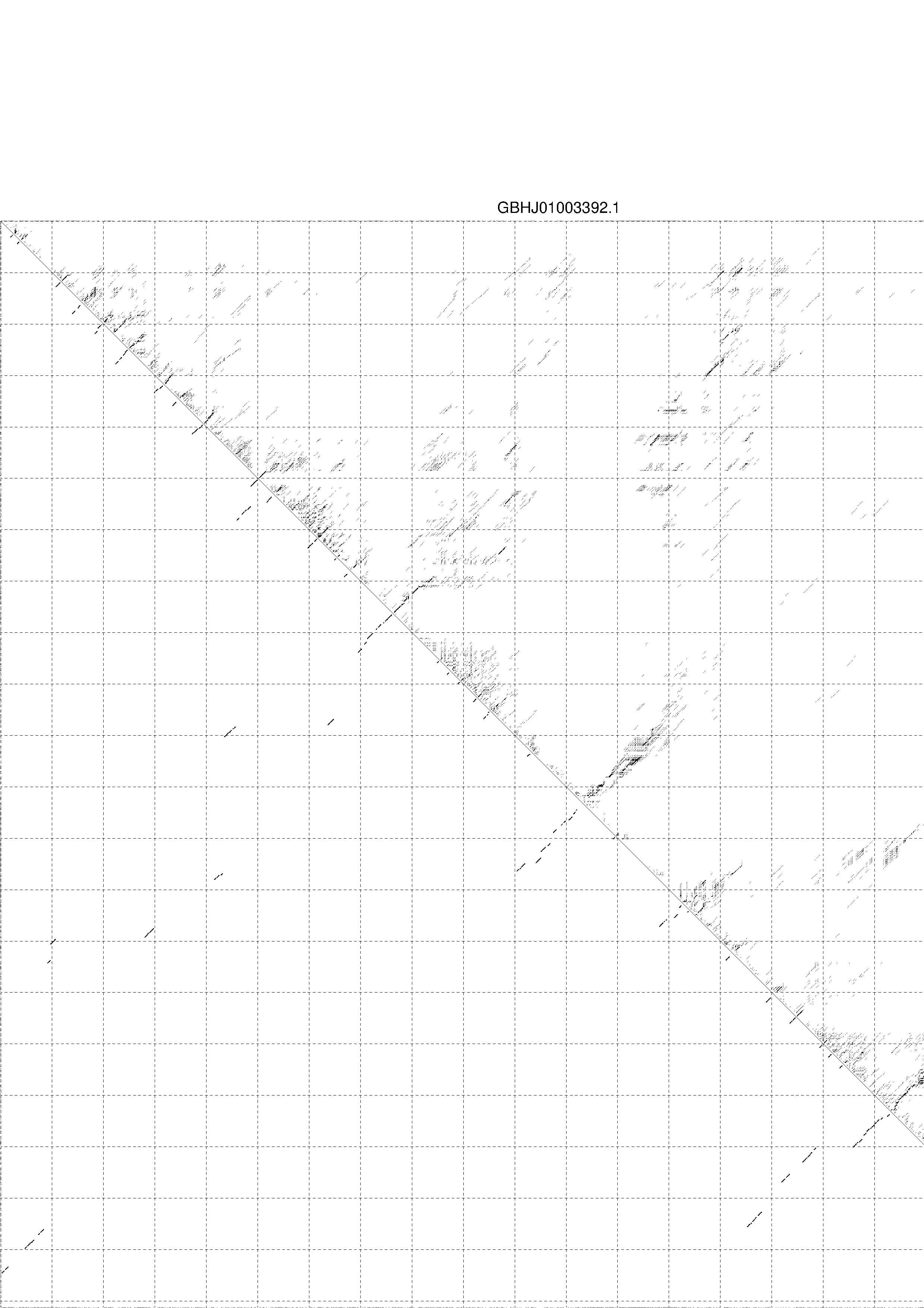
4. You may look at the dot plot containing the base pair probabilities [below]
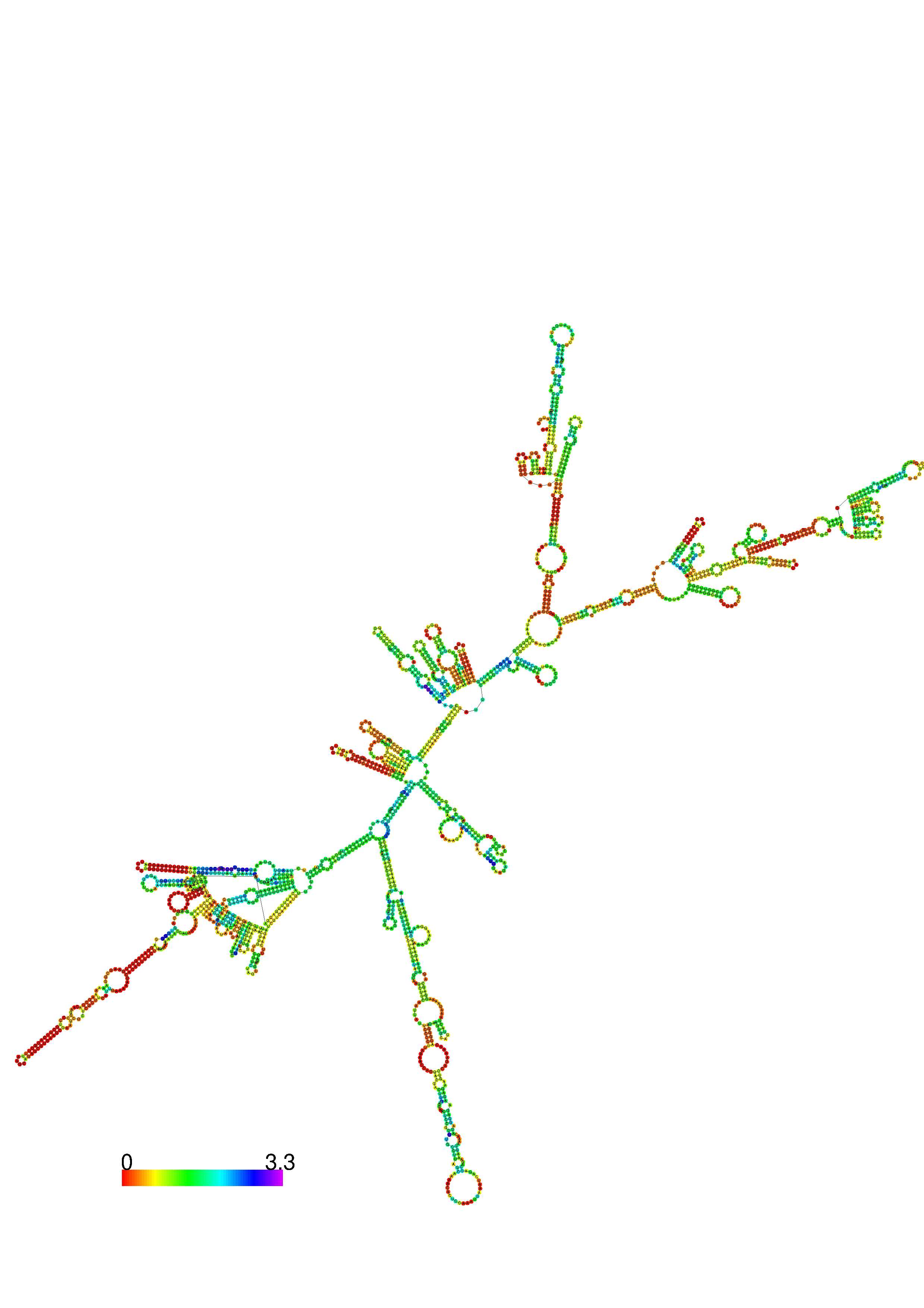
III. Graphical output
1. A note on base-pairing probabilities:--
The structure below is colored by base-pairing probabilities. For unpaired regions the color denotes the probability of being unpaired. The reliability information of RNA secondary structure
plot in the form of color annotation, Positional entropy ranging from red (low entropy, well-defined) via green to blue and
violet (high entropy, ill-defined).
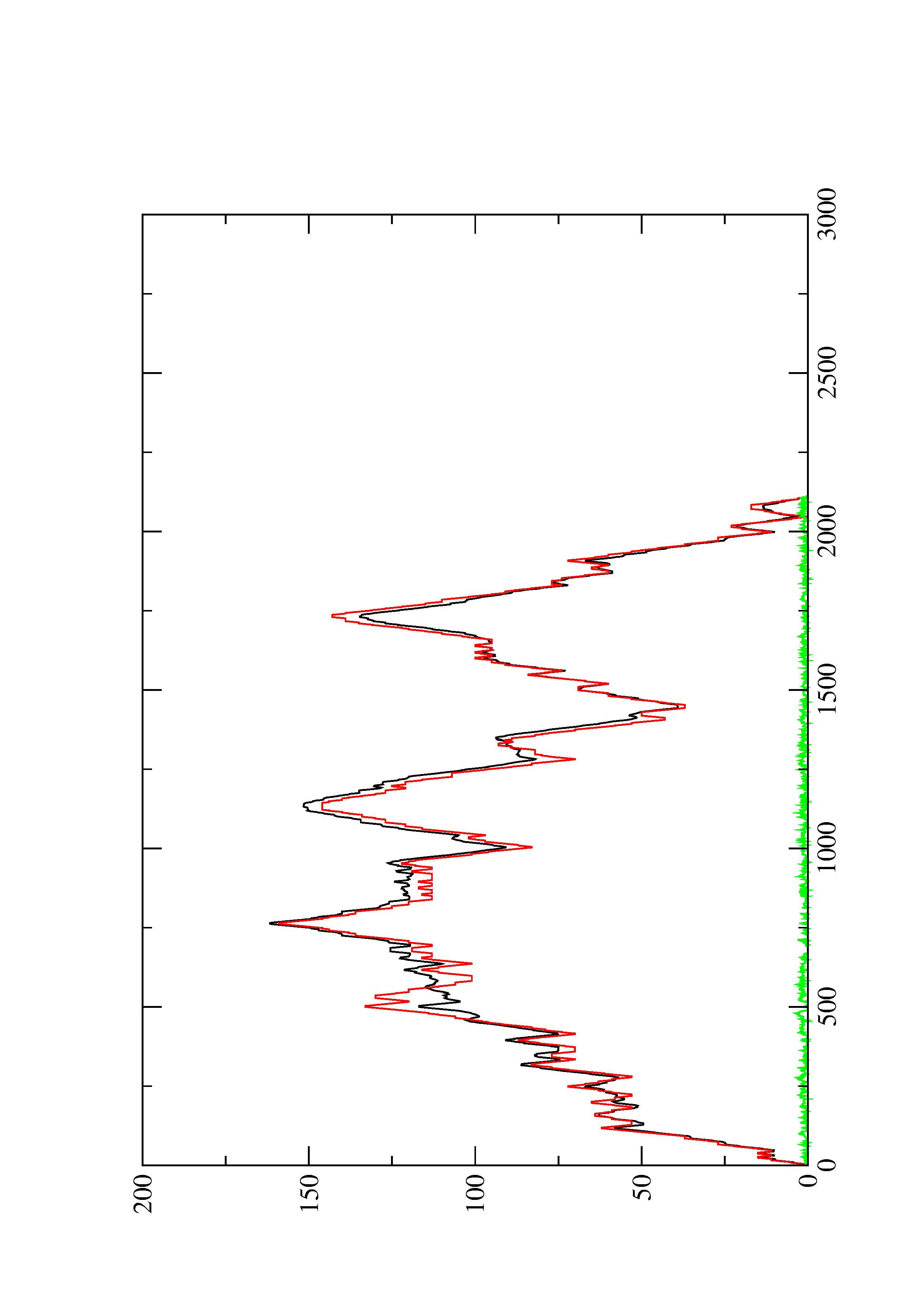
2. Here you find a mountain plot representation of the MFE structure, the thermodynamic ensemble of RNA structures, and the centroid structure. Additionally we present the positional entropy for each position. The resulting plot shows three curves, two mountain plots derived from the MFE structure (red) and the pairing probabilities (black) and a positional entropy curve (green). Well-defined regions are identified by low entropy.