lncRNA Structure Prediction
Species Name : Withania somnifera Sequence Id : GBHJ01003868.1
I. Result for minimum free energy prediction
The optimal secondary structure in dot-bracket notation with a minimum free energy of (-569.10) is given below.
1. Sequence-
UCUUCUACCCCCAAACAUAGCCAAAAACCAAAAAAAAUUGUGUAUGGAACUAUUUGAUUU
UAGGGUAUAUAAUUGAUAGGAAGAAAGGGUGGUGAUAAUUGUUGACAGAAGAUAGAGAGC
ACUAAGGAUGAUAUGCUAAAUUCAAAUCAAUUUUAGGGAAAAAUUAAAGCAUCUCAAUUC
AUUUGUGCUCCCUAUGCUUCUGUCAUCACCGUUGCCCUCUUUCCUCUUUUCCUCUCUUUC
AGUCCAAUUAAUUCAAAAUCGCCUUGUGUAGGGCUGCUCUCCCUAUCCAUAGUUGUUUUU
UUAUAUUCACAAGAUCUCUCUGGUUAAGGAUUAGCAAGCCCAUCCACUGCCCGCAUCUUU
UGUUUUGGUGUAUGUAUUUCACGUACAAAUACUGGAUGUUACUUUCCACAUGACAAUUUU
UUAUAUAUUAUACUAUAUAUAUUACUCAGUUUAGGUAUUAUUUGGACUAAAGGAUAGAGU
UUUAGUUCUAAGGGGUCAUGAAUUUUUAUACUUGAAGACUAUUUUGGGUUUUGCAAUUUU
GUCGAUUGGAAUUAAAAUUAGAUCUGAAAGUGGGAGAAAUAUUAUUAGUUUGUUCAAUUA
CUAACAUUAUUUUGUGAGUUUUAUGUAUAUUCCUUUGCAGUUAUUGGUAGAAGAUCCAAA
UCUCAUGGAUUUCGCCAAGCAUUCAACGGAGGAUACGUCUGAGGAAUGAGAAUUUAUCAU
UUAUCCAUGUAAUUAACCCUCUACUCUUCCUCUUAAUUAAUUUGAAUGGUAGUUUACAAU
CUAAGUUGGUUUGGAUAAUGUAAUUUUUCUUGGAGCUCAUUAAAUGGUUUACACAAAAUA
GAUUGAACUUUUACUUAUUUUACUUAAUCAAUUAGACAGAGGUAAUUUUAUUAAAGGCUG
GACUUUAACUGUGCUUAUGCGCACACAUCUUUUGCCCCCCAUUUUUUUUUUUUUUUUACU
UUACGGCUACACUAUGUUGCUUACUCUUCAAAAUUUUUUUUUUUUUUAGUACUGACAUGC
AUCUAUCAAUAUUUUUUAAGUCUUACAUUAGCGUAUGGCAUAUGAUAAUAGACUUUGUAA
GAAUAGAAGAUCAUGUGAUGGUUAGGAUUUAAUGGAUUCACAUGUUACACCAAACAGAAA
AUAUGGUGUGGUAAAUGAAUAACCGAUCGAUCACAACUAGGUUAUAUAUAUGCAUAAAAA
UACUCAGCUCCUUCAUUAAAAGAAAAAAAAAGACUUAAAUCUCUGGAUAAUUAGUAAAUA
AAAAAGAUAAGGAGCUAGGUGCACAUGAUAAUUUUAUGGUUAUCAUCACAUGUAAAUUAC
UAUUUAAUCCUUCUUUAGAGGGAAUUGUUCAAGAUAGGUAGAAGAUUUUUUUUAAAAAAA
AAAAAAAACUGAAGAAAGAAGGUUAAAGAGAAGAAGACUUCAACAUAUUUUUGCAUCGAA
AAGUUAAAAAACAAAGGUCAAAAUCUAUAUAUAAAUUAUGUCUCUUCUGCUCCUAAUUAG
AGAUAAGUAGUAAUUAAUGUCAAAGUAAGCUUAAAGUCAAUUGGUAUGACUUAAAGUAUG
AUUUAGUUGGCCAAGGCAAUUGGUAUGAUUGACCAAUUCUCUAGAAGAUAUCAAUUGUUU
GUAGAAGUGUCUUAUACAUUUAUGGGGAUGAAAGAUAUGUUGUUAUUUCUCCUCUUAUUC
UAAUUAAACGUUAGAGAUGUGAGAUUAUAUUAUGUAAGUAGCGAAGAUGUACAUUAUUUA
AAUUUUGUGUGAGCUAAGUAAAUAUUAGAUGAAAAAAAAGGUCAUUUCUUAAUCUGUUAU
UGGACUGGAAUAUCACUAGUACAGUGGAAAGUGCAAUACUAUAUGGAUGUCUCUCAACUU
GGUCUAUAUGUGUUGUAAUGAAGUUAAAAAGUGUAGUAGUAAUAAUUUUGAACAUUCCCA
AGACAUGCAAUACACAGUGGCAGAGCUAAAAGUUCUGAUGAAAAAAUAUAUAUUUCCAAG
GAGUCUAAUAAGUGGCACCAAAUAUAGAAGGAUUUGUGCUUUGAUGUGAAUGCCUCUUGU
UGUUGUAGCUAUAGUAAGGGACAAAUGAAGUGCAUUUUAAUAAGAAUUCUUGGAUGGAGC
AUUUCAAUUGAUGAUCUAAGUUUGACCAAUAUGCUUAAAGAAGGGGGACACGAGCAACCU
CGAGUUGAAGAAGGUAAUUCAUAGAAUGUAAAGAGAAUGUUGAGGUUGUCAUAUCAUAUU
GUUUUAAAUUAGGGAAGGUAAUCAGAUAGCUGAUGUCUAAGCUAAACUCAGGACCAUUUC
AACUAGAUUGGUGAUAGCUUUCUAGUACAUGACAAGAUAUUACAAGGUCAACUUUUUUCU
GUUUGGGUAAGUUAAUGAUAUACAUAUGAAAGUGACCAUGUAUAGAAAUAUCUCUUAGUC
UUUAUCUUUUGGGAGAUAAGUCUCAUGUCCCCUUCUUCUCUUUACUACACCAAAACAUAU
AUUCAGCGGCAAUUAAUUAAUGACACUUAAUUGUUGCUAAAAAAUAUUUUUAAGGGC
2. MFE structure-
.......((((((.((((((.................)))))).))).......(((((((.((((...((((((((((((((((((((.((((((...((.(((((((((((((.((((((.((((((((.(((((.((((....((.......))....))))..))))).)).)))).)).)))))).)))).))))
))))).))..))))))))))))).....)))))........)).)))))))))))))))))(((((....((((((((.(((.((.(((.((..(((((...........)))))..)).))).)).)))..))).)))))..((((((...(((.....(((((((((((((((.....)))))))......(((((((
((((((((((((((..((((((.....((((.(((.((((.(((((.(((((((((...)))))))))...((((..((((((((((((((..((.((.(((((.....(((((((......))))))).....)))))))))..))))))))))))))..))))...(((..((((((.(((((....((((((((..(
(((((.(((..(((((((....((((((.((((.(((.(((..(((((...(((((((.......))))))).)))))..)))))).)))).))))))..((((((.(((..........((((....))))...........)))))))))((((.....)))).......)))))))..))))))))))))))))).)
)))).))))))...))).......(((((((((((((((.((((((.................))))))(((((((.(((((........((((((......))))))..((((....))))(((.((((((((..((((((....(((.((((((.(((((.((((.....(((((((.....(((((......(((((
((((((((((....(((((.((((.((..(((((((.((((((..(((..((.....))..))).....))))))...)))))))))))))))))).......((((((.((((((((.(((((((((((((...........)))))))(((.(((.((((((...............)))))).))).))).......
((((.((((((((.(((...............(((...(((....)))....)))..........))))))))))).))))...(((((((((....))))))))).)))))).)))..))))).))))))..(((((((((((((.(((..........))))))))))))))))...........)))))))))))))
)).......)))))......))))))).....))..)).))))).)))))).)))((((((((((...((((.((((((..((..(((............)))..))..)))))).))))........((((.((((((.......)))))).))))..))))...)))))).(((((((((((((..............
.......)))))))))))))....((((((.....)))))))))))).)))))))).)))..........)))))...)))))))...(((((...((((((.......))))))...))))).(((((.....)))))..)))))))))))))))))))))))))))))))))))))..)))))...............
((.(((((.......))))).)))))))))))(((((((.((((((((.............)))))))))))))))...(((((((................))))))).))))))))))))))))))......))))))(((((((...)))))))...................)))))....(((((((.((((...
.((((.((((........)))).))))...)))).))))))).........(((((((((.(((.((((((((.(((...((((....))))...))))))))))).)))......(((((...........)))))(((((((((.(((.((((((((((((.............((((...))))............)
))))))))).(((((...(((((.........(((((((.(((...(((((........)))))........((((..((.....)).))))))).))))))).......))))))))))((((.(((((.(((((...(((...................)))...)))))))))).))))..((..((((((((((..
........))))))))))..)))).)))))))))))).))))))))).................((((((((((((..........))))))))))))...............))).
You can download the minimum free energy (MFE) structure in
II. Results for thermodynamic ensemble prediction
1. The free energy of the thermodynamic ensemble is [-619.17] kcal/mol.
2. The frequency of mfe structure in ensemble 5.25324e-36.
3. The ensemble diversity 600.67.
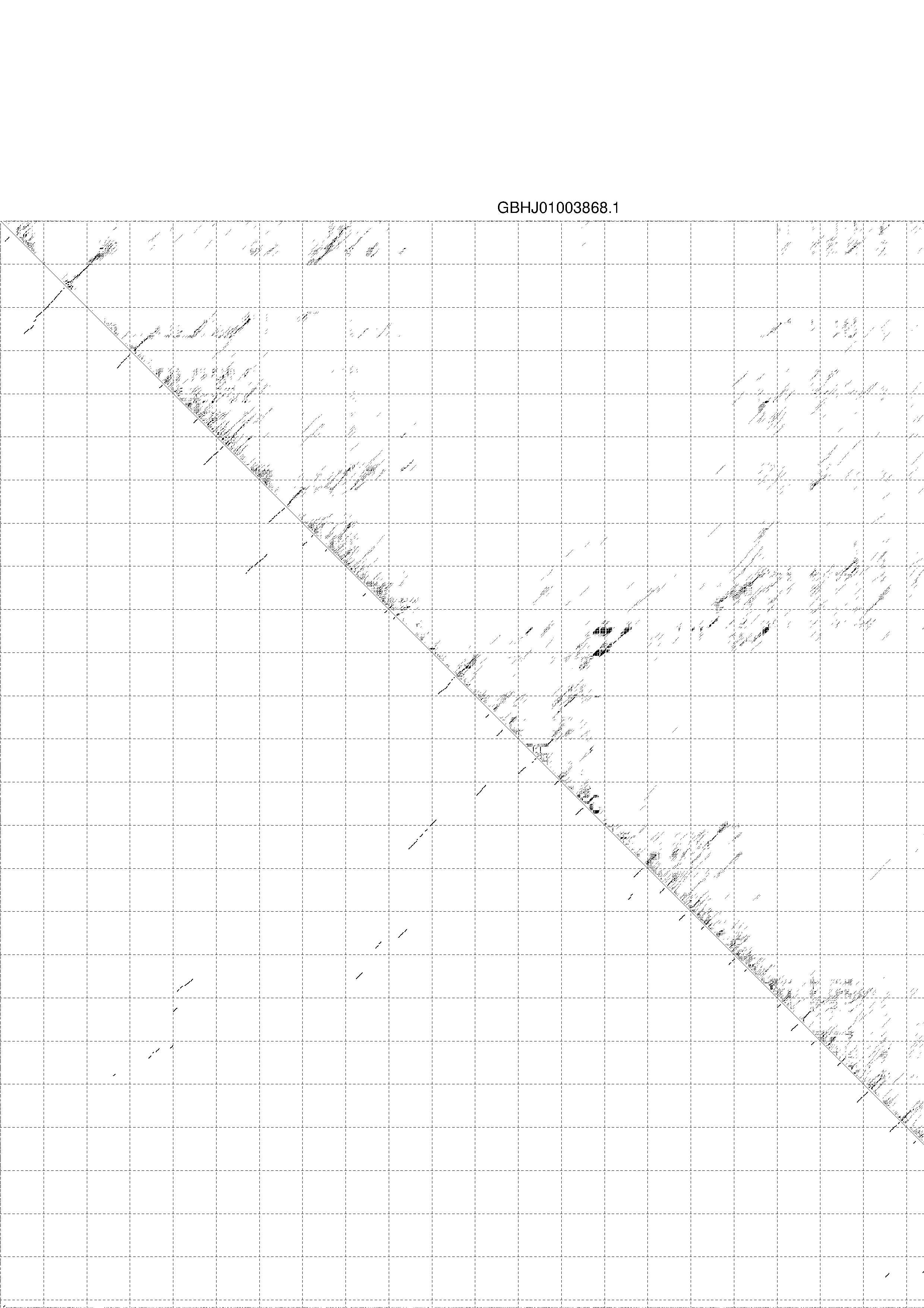
4. You may look at the dot plot containing the base pair probabilities [below]
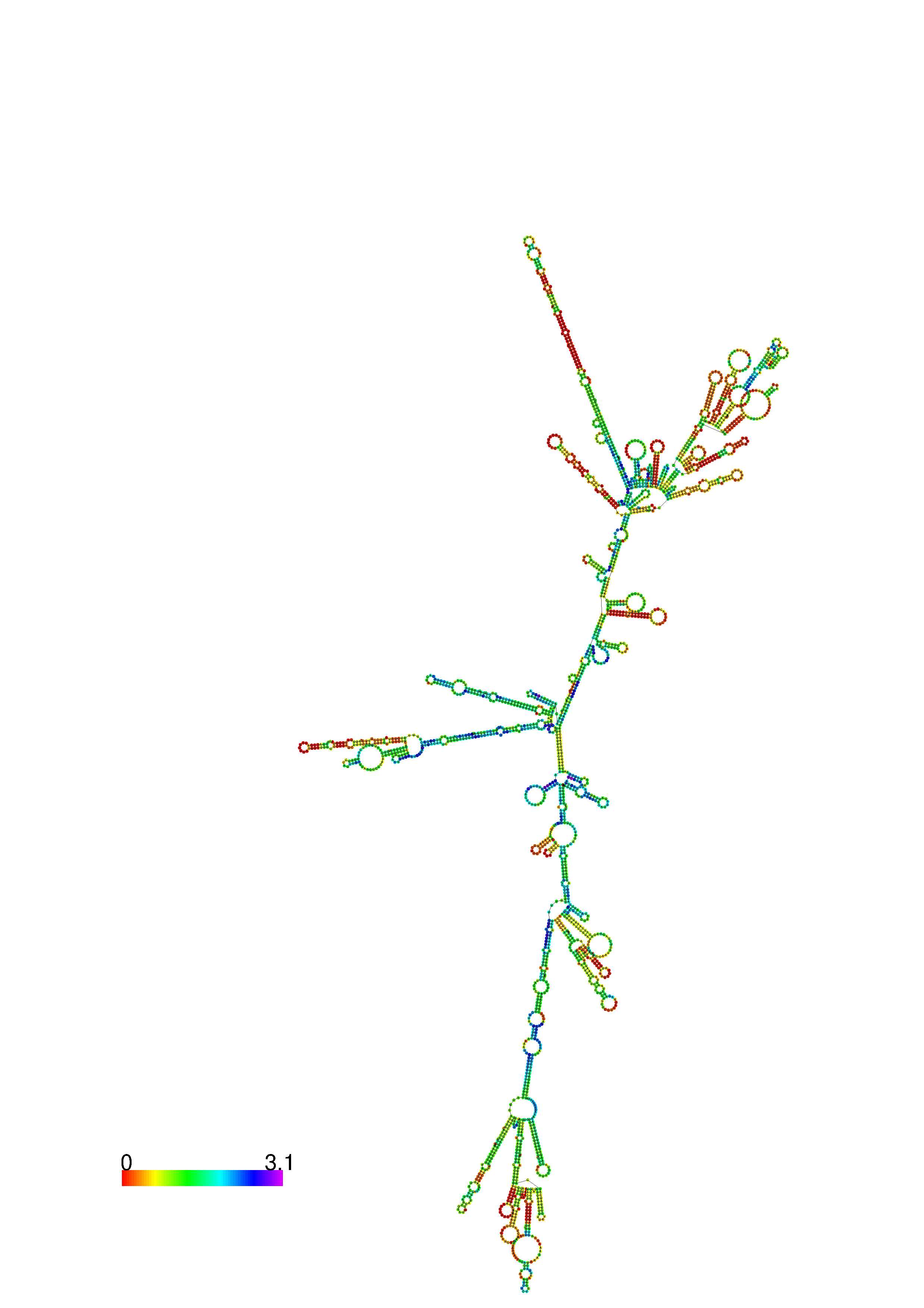
III. Graphical output
1. A note on base-pairing probabilities:--
The structure below is colored by base-pairing probabilities. For unpaired regions the color denotes the probability of being unpaired. The reliability information of RNA secondary structure
plot in the form of color annotation, Positional entropy ranging from red (low entropy, well-defined) via green to blue and
violet (high entropy, ill-defined).
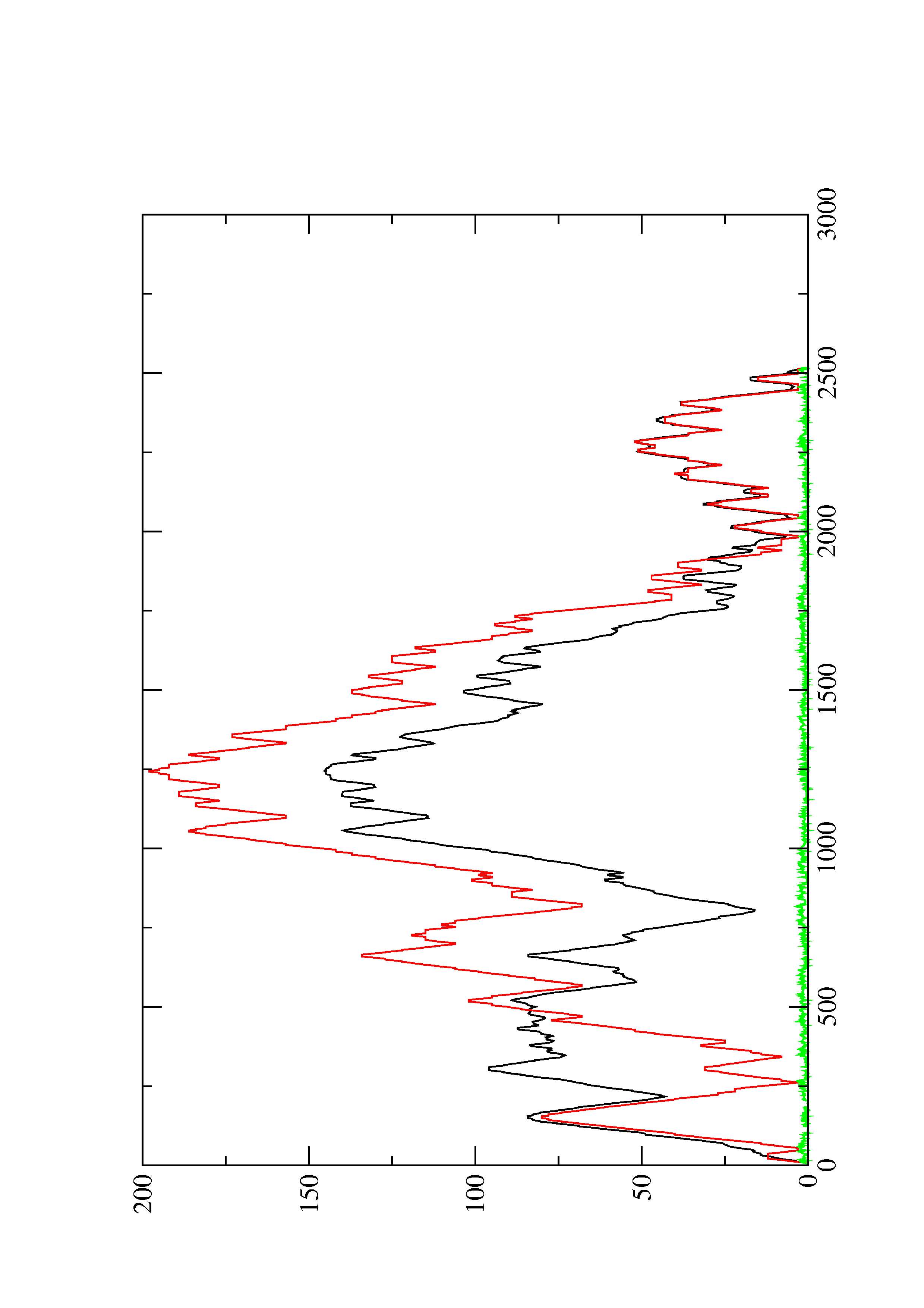
2. Here you find a mountain plot representation of the MFE structure, the thermodynamic ensemble of RNA structures, and the centroid structure. Additionally we present the positional entropy for each position. The resulting plot shows three curves, two mountain plots derived from the MFE structure (red) and the pairing probabilities (black) and a positional entropy curve (green). Well-defined regions are identified by low entropy.