lncRNA Structure Prediction
Species Name : Withania somnifera Sequence Id : GBHJ01004769.1
I. Result for minimum free energy prediction
The optimal secondary structure in dot-bracket notation with a minimum free energy of (-625.60) is given below.
1. Sequence-
AAACCUCAGCCGCCAUUCUAUUCACUAUGCUUUCUCCUCUCUCUAAAAUAAUACACAUCC
CUCUAUCUAACUCUCUCUCUCUUUCCCCCAACUCCGGCCGAUAACCACUAAUACUAGUCG
GCCCACUAUCUCCUCUCCCCUCUUUCGUUCACUCUCUUCGCUGAUCAGCAGCCAUCGAUG
GCCGGGAAAACCCAUCAGCCUCCACCCUAAUCGCCUCGCCGUUCACUGAUCUCGACUUAU
AGAAAGAGAUACUUGUUAGACAUUUAUUGAAGGAAUUUCUUGUUCUUUCUGGGCUUUUUU
CUUUUUGGAAACUUUUAGCUCAUUUUGGACUUAGACGAGCUCGCUUUAUCACAUAUUCUU
AGUUAUUCAGGGUAUAUCUUGAAGUUCUUUUUAUUUUUCCUGAUUGAGAUGAGCUUGCGG
AAGGUAUGAAUAUUUUUUCACAUCUUCUUUGGUGUGAUGCCAACCUUUUGGAUGAUACUA
UGUUUUGGUUUUAAUUUAUUUUUUAAAGUAAAAUAUUGUGCUAGUGUUAUAAUUUUCACU
GAUUAAUUUUUUCUGUUCUGGAUUCAAAAUGAGCUUUUAGAAGACUAAUCAAUCGAGAUU
CUCAGUUGUGUUAAUAUAUAUGUUGUUGGUAUUAAUUGUGGAGUAUAUUUCACUUAUAGC
UGACACUGGAUGAAUAAACUUGAGACUGAAGUUGAUUCUUGAAUCAACUAUUGAUCUUGA
AUUUUUUUUUAAAGAAAACUGUUGAUCUUGAAUAAUAGUGUGUAGAUAAAAUUGAGUCUC
CCACUCAUUCAAAAAUGCCCUGAAGUGAGUUGAUGUCAAAACCAUGAUCAGUACAAAUAG
CUCUAUUUUUUAGCUACUGUUUUUCAGUUUGUUAAACUAUAGUGAAGUAAGAUCUUAAGA
GUUGCAUCUUUCUUUAUUGUGAGUGAAACAAUCAUCUUCAAUUUGGCAAUGAAAUUUUCU
AUGAGAGUUUUGCUCUAUCUAUGGCAUUUCUAAUCCUCAAUAUUUUGUUAUGGAAUUUAU
AUUGAGGAAUAAGACUCAUAUUUUCGAAGAUUUGUGCCCAUUCAAUUUACUAUUUGAAUU
GUGGCUAAGGACUUUGGUAAGAAGCAGGAGUUCUGGUUGAUCUACUUAGAUGCUAAGGAC
AUAUAGUAUCAUACUAAGUCUAGUUUCUCCUAAUAAUGUUGUUAUUAUUGAACCUUUCAA
UUACUUCAAAGUUUCUUUUUUUUAUGCUGUUGUACAACUCUACUAGUCAUGCAAUUGAGA
AAAACAUAACUGAAUCUACUAACUGCAUUUUCAUAAACAUCUGUACAUCUCUGGAUGAGC
AUCUUUGCUAGUUACUAGAGAGCUUGGAAUGUAUUUAUAAUUAUCCAAACAUCAGUAAAU
CUGCAUUCAAAAAAUUUUGUGCGAAAUGCUAGGUAAAGUUGAUAAUCAUAGCAAUAUCCG
AAAAACAUAAAUACAUACAGAGCUUUGACACAUGGAUUCAUUAUCUAUAACUUAUGAAGC
AACUGUUAACUUUACAGGUUGACUAGUGGGAAGUGUAUAUGAUUUUCACUGUAGGAUCUU
GAAGUUAUAACUGGGUGUAAUUUUGAGACUGGGAAGGGGGAGACAUUGAUAUUGAUGUCU
UUAUACAUUUGGAUUUUGUAUGAAUAACUCCAUUGAGGUAUUUCGAUCUGACCGGCCUAU
ACUUUAGAGUUGGAAGCUGAAUUCGUGAAACAUAAAGAAAUUAAAAAGAGUUGCAGGAAA
AGCAAGUAUGUUGUCAUUGAAAUUCACAGCACCACUAGAAGAAAUUUAUUGUAAUAACUG
CUUGCUUUUAAAUGUUUUUUGCAGGCUGAAUUUAUUGACAUGCAGAAAAAUCAGUUAGUA
GCCUCUUUUAUGACUAACAAGUAUCUCUUUCAGCCAGACAACAAUAAUGGUCAGGGGAUU
UGGUAAUGCUUUAUGGUUUGUGGGACGCUCUAUUGAAUAUGAAACUACUGAUGAGUGGAA
GGUGUUGUCUUUUAAUACCACUGACUUGACUAGAGUCUUUCUUUCUAUAGCCUUCUUCCU
CAAUCAAAAUAUUAAAAGGAUAGGCUGAAUUUACAUGUUUGCCUAACCAUACAUAUCUAU
GAACUUUCACUUUCUUUAAUUGAAGAGACUGCCGGAGUAUGUAAGUGAGGCUUGGAACUU
UCUUCCUUAGUUUAAUGUGGAUUCCUUGGUUGUGUUAGUUGUUCUUGUGGUCAUUGUUUA
UGACUCACUGACUGUCGCCCUUUGUAAUGAUCAAAUCAGCUAAUGAUCCAUAAUAACCUG
AAUCAUAUAAAUGAAAAGAAGUAUCUGAAGACCCAUUUUCUUCUUGUUGAUAAACACUGU
GGUUAAUGAUUCAAGCAGCCCCAGGGAUUUUAUUUUGUUCGUGUUGGGUUUUUAGUAUGA
UUGGGGUUUGAUCCUAUGACCUUUGGGUUAAGCUAAUGGAUGGAAAGUGAAGGGACUUUG
UCCCACAUAGGAUGAAGAAAAGAAAUUUUGUUGGGUAUAACAUAAAGCAUUGGGAAGAUG
GUAUGACCUCGCGACAUAUCUCUGUUUGGAUUGGCUCGGUGGCACACCUAAAUUCUUAAC
AGAUACCUCCUAUUGGCUAUAAAGUCAGAGUUUUAUACUGAAAUUUGAAAUCCUUUAC
2. MFE structure-
........((((((.(((((((((.((.((...........................((((.(((((............................(((((((..............)))))))....................................((((.....))))....)))))..))))...(((((.((((
...(((..((((.(((.((((((...((.(((...((....((((((((((((((((((.........((((((((.....))))))))((((((..(((((....))))).....))))))..((((((((((..(((((((((((((((....(((((((((((...(((((((((((.((((((.(((((.((....
.)).))))).))))))...((((((((((........(((((((......)))))))..((((....))))(((((((((.((....((((((.((((.....)))).)))))).....))))))))))).........((((((...(((((((....((.((((...))))))..))))))).)))))).((((((.(
(((((((..(((.((.(((....(((((((..(((..((((((....)))))))))..)))))))....))).)).)))....))))))....)).))))))((((((..((..(((((((......))).)))).))..)))))).((((((...((((...((((........))))..)))).))))))((((((((
(((.(((.(((........))))))..))))...(((((....)))))((((((.((((.....))))..))))))..(((((((((.(((((............))))).)))))))))(((((((((..((((....(((((.......)))))....))))..))))))))).......)))))))....(((((((
(((...............)))))))))).......)))))))))).)))))..))))))((((((........)))))).)))))))))))...))))..)))))..)))))).(((......)))..(((((((........)))))))...))))))))))........(((((((....)))))))(((.((((.((
....)).)))).)))(((((((((....((((((..(((.....)))..)))))))))))))))(((((..((..((((((((((.(((((........((((((..(((((((...((((....)))).....)))))))..(((((...............)))))...((((((((((.((...((((((((.....
.(((((((((.(((..(((((....(((((.(((.(((((((((.((((.((((.......(((((.....((((((.....)))))).......))))).(((((((((((...)))))))).))).....)))).)))).))))).((...((.(((((((((((((......)))))))))))))))....))..((
(((((((......)))))))))........)))).)))))))).)))))..)))..)))))))))......((((((((.....))).)))))...((((((((.......................))))).))).(((((((((((.(((((((.((((.(((...............))).)))).))..)))))))
)))))))))....))))))))...)).)).))))))))..)))))))))))))))))))))..))..))))).))))))))))))))))))))..))))).....)))))).))).)))).)))..........)))).)))))..))..)).)))))).))).(((((....)))))..(((((((.....))))))).
..(((((.....)))))...........((((....(((..................((.(((((.((((......))))))))).))...................((.(((((((....))))))).)).((((((((((..((((((.(((..(((.(((((((..((((.((((....(((.(((....(((((((
(......(((((((((....((((.........)))).......)))))))))...))))))))...)))....(((((.(((((((.................(((((((((((.........((((....))))(((.((((............)))).)))((((........))))....))))))))))))))))
)).)))))..((((((((.((.((((......)))).))............(((((...))))).)))))))).((.((((....)))).)))))....)))).))))...))))))).))).))))))))).))))).)))))...)))..)))).))))))................((((((.(((....(((((..
..))))))))...))).)))..................
You can download the minimum free energy (MFE) structure in
II. Results for thermodynamic ensemble prediction
1. The free energy of the thermodynamic ensemble is [-671.80] kcal/mol.
2. The frequency of mfe structure in ensemble 2.76315e-33.
3. The ensemble diversity 732.36.
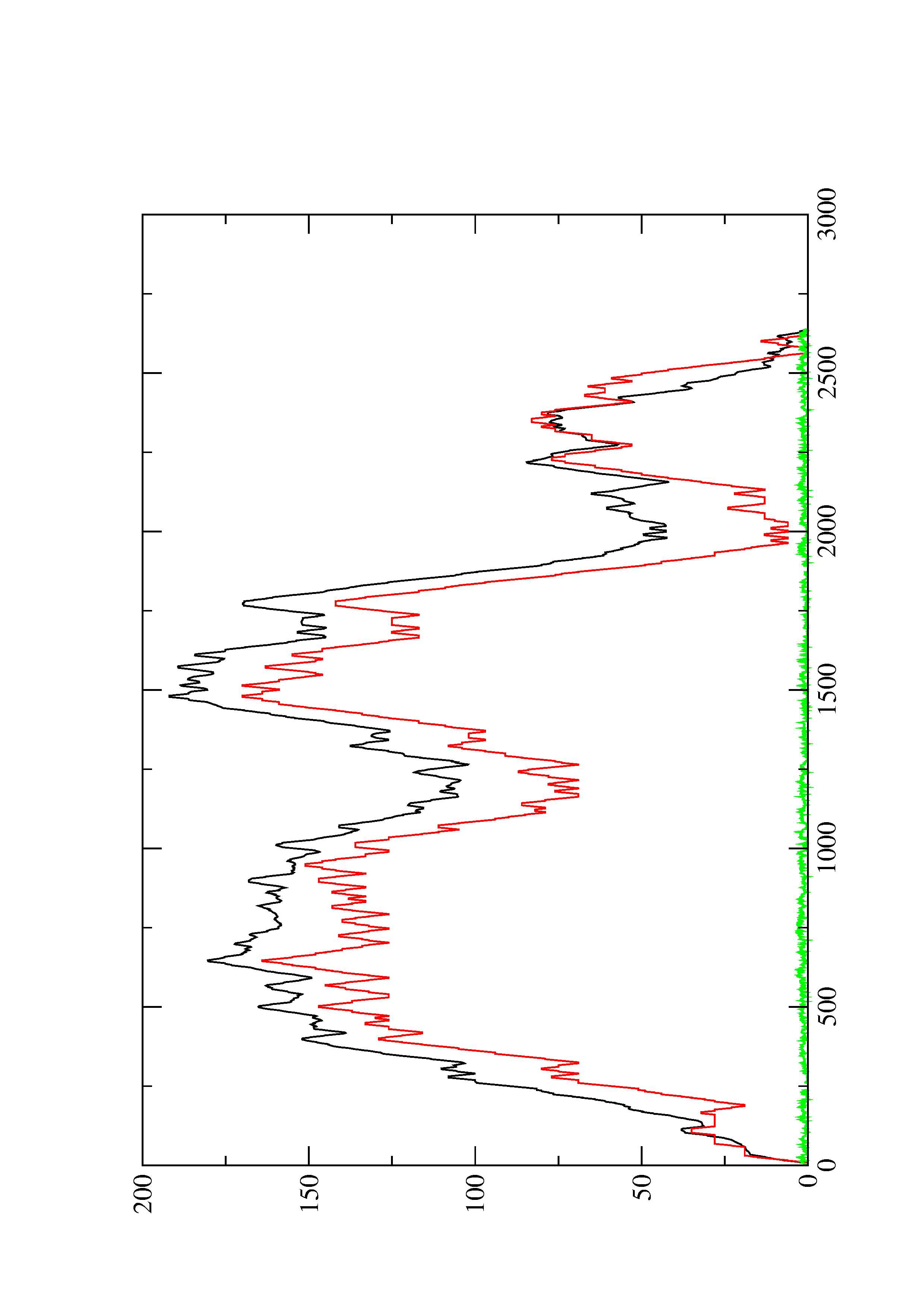
4. You may look at the dot plot containing the base pair probabilities [below]
III. Graphical output
1. A note on base-pairing probabilities:--
The structure below is colored by base-pairing probabilities. For unpaired regions the color denotes the probability of being unpaired. The reliability information of RNA secondary structure
plot in the form of color annotation, Positional entropy ranging from red (low entropy, well-defined) via green to blue and
violet (high entropy, ill-defined).
2. Here you find a mountain plot representation of the MFE structure, the thermodynamic ensemble of RNA structures, and the centroid structure. Additionally we present the positional entropy for each position. The resulting plot shows three curves, two mountain plots derived from the MFE structure (red) and the pairing probabilities (black) and a positional entropy curve (green). Well-defined regions are identified by low entropy.