lncRNA Structure Prediction
Species Name : Withania somnifera Sequence Id : GBHJ01005148.1
I. Result for minimum free energy prediction
The optimal secondary structure in dot-bracket notation with a minimum free energy of (-543.80) is given below.
1. Sequence-
CUUUUUCUUCUUCGACAUCAACCAACACAGCUCCUUUGUCAAACCUCAAACCCCCUUCUU
CUUCCUCACUUUCUCACCCUAAAGUCAAAGAACCCCAAACAAGCAACCAGAAGAAGCUAA
CCAACACCCCUAUUACCCCCAUAAGGCUCCAUUUUUUUUCUCCAUCUGUGACAUCUUGAG
CAUCCAUUAUUGACUUUUUAUGGGUGAAAUAUUUUAUUUUUCUUGCAAAAAAUCUGAUUC
AAAUCAGAGGCCAUCAGUUGAAUCCGGAUUUUGGGUAAUUCAAAUCUCUAUUUUCUGCUA
UAAAUAGAUGUGGUUGUGGCCAUUUGAAGGGGCAUGGUUUUUGAAGUUUCAUACUCCUUC
AUUGCUAACAAAGGUUUUUCAAGUAGCAAUCUCUCCAUAAUUAACUGAGAAAUGUCAUAG
UUCUUAACCAUCAACUAUUCUCGAAAGCAAAAGUUAAUUUUCUUUUCGUUUGUUUUCUCA
CUGAAAAUCCAAGUGGUGAAAGCUCGUUCAAUUGUUCCUAGACUCGUUGUCUUUGUUCGC
UGCCCAAAAGAGGUAAAUCCGACUCCACUUUACUCUGUUCUCUGCUAAACCACAGUAUUU
UUGAAAUUAUUCCUUAAGAUCCUUACAUUAUUUAUGUUCCUAGUUUUGUGUCUGUUGUGA
AACCUAGACAGAAUGACAUCUAAAAGAUGUGUUUGUUUCUGCUAUCUUAUUAGAAUGUGA
CUACGAAGUUAUACUGUUUUGCAGAAAUCACGUCACUUGGGACUACUCUUAAGUUUCAAU
AUUGACAUUAGUAGCAGGGCCCUUUCUCUUGUGUGGGGGCUUUGAAUGAAAUUUAAAUAA
UAAAAAAAAAAGUUUAGUCUCAACAGACUACGAUUUUGAUUCCCUUACUAAAUAUGCAGU
AGAGUACAUAACUUAAAUCUCUUUCUUCAUAUGUUAGAUAUCAUAUAUUUGGAAAUAUAA
UUUUUAGCUAAGUAUGACUUGAAAUGUUGGUGUCUUAUCUGCACUUUAAUUGAAUUUGGU
UGUGCACAUAAAUGUUAGCAGGUGUCUCUCUGUUAGAACAUUAGUUUGUUCUCACUUUUC
UUUGUGCAAGGAACCUGUUCCUUCCCCAUUUGUAGUACCGUUUCUUUUACUUUUUUCUUC
AAUUUCUUUAGCCUAUACAUGCUAUAUCUUUAAGUUAGCUGUAGAAUUUAAUUCGUUUUG
UGUCAAUAUCGUUGUUGUUGUCAAUCUUUUACUGUAAGCAAUGAGUUGGAAUUAUUGUCU
AGAAUUAACCAAUGAUUUGUUGAAGUCCUAAAAGUUGUUUCACACCUUCGGCUUGAGAGU
GCCAACUUCCGACUCGUCCAUUUUUUCUUGCUAUUUGUUCUUUUCUUUAAAGUUCUUUAG
UGCCGCAAAGCCAAAGGUAUUCAUUGAUCCCCGAAGUUUUAUUUUAAAAGAAAGAAACCG
CUUGUGUAUUUAAGUUCUCUUCCUGUGCACCUUUACGGAAGUAGGUUGUCAAAAAAUACU
GUGUAUAAAAUCACUUAGAAGUACUUGGAAUGAACAUAAUAAUACGUUUGUUUUUACUAG
CUUAUCAUUUCUUUGUUGCUGUCAAUGCUAGGUCUAGCCUAGUUUCAUCUCAAAUGUUCU
AAACUCAGGCCUUUUAGUUCUUCUCGGUUAUUUCUCAUGGUAGUCAUCAGUUCAUGCCCC
UUUCCUAUCCCAAAUUACUAAAGAUUCCCAUAUCCCAUGUCUUUGCGUGUUUGAAUCUAA
UGACAUUUUGCUGUUGGAUUCAGAUUUUUUAGUUAAAAAACAAACAGGUCGUGACAAGGA
UUUGAGCGAGAAUUUGAAUCUGAGCAUGUUUUUAGUUCUCAGCAAACAUGUCAAUGUUAU
CUCUAAGCCUACACCUUAAUCUAUAAUUGAAUCUCGACCUAUUUAAUAAGGUGUUGAGUU
AGUAUGACUUGAAUCUGCAAACACCUGUUGCUUAGUCGCUGUUAAUACUUGAUUUAAACA
UUUCUUAUUAUUUUCAUCGAAGAUAAGGGGAGGUAUAAAAGGGAACUUGUUAAAUACUAG
UUUCUUAGUAAGUUCUUGUUGCUCAAGUUUCAUAAAACCUUUGUUCAACUUCAAUAAUCU
UUGUCAAGUUUAAAUGUAGUAAGUUAAAAUUUCUGUAUGUGCACGUGUCUUGUCUUUUGU
CUUGUUUUAUCAUUUUUCACAUGUCUUUGGCUUUCAAAUUGAGCAUUAGCACGUACAGUU
GAGUUCAUUACCUAGUUUUAAAUUUAACUUUAUGUAUCCAAAUUUAUCUUAGUUACUCAU
GUGUUUUCUAACUUUUCAGUUUGUUUUGUGCAUGGGUACACCAAAUGAGUCCGACGGACU
CGUCAUAUGUGCAUUCUCUCCGAAUCAUCCAUUUAGCCCAUUUCUUAUGAUCAACCGAAA
AGAAAGAGAAAUGGGUCCCCAAAAUACAUAUUUCGCUGUGCUUUUUCAAAGACCCACUUC
UUGUCGGACGAUCAGAAAAGGCAGUGAAAAGAUCAAAGCUAUUAGAUGUUGGGUCAAAAG
CCUAAAGUCAUUUUCCUAUCCUUAGUACUUCUCUAAAUUUUUCAUUUUAUGUAUGCUUUG
UAACACUAACAAUUUUUAUUUUCUUGUUCAUUUAUUUUCUUUAUCUAGGUGAAUCCUCGG
ACGAGUUUAUUUUAUCCAAGCUUGGUUUCCU
2. MFE structure-
(((((....((((((((....(((((.......(((((..(((((......(((((((...............((((((.((((((((...........((((....(((((((((...........((.(((.......))).))..........)))))...))))......))))...........))))))))...
.))))))...............((((..((((((((((((((....((.....))..)))))).))))))))..))))........(((((((........))))))).(((((....)))))..)))))))...))))).)))))............((((((((.(((((((((...(((.((((((((((.......
......))))......((((((..........))))))...((((((.((((.....)))).))))))...(((((((((((.........))))).))))))((((((((........((((.....))))......(((.((((.((((((((((...........)))))...((((((((((((....((((((..
.(((((......(((((((................(((((.(((..((((....(((((((((.(((.(((((..((((((...))))))......))))))))(((....))).(((((((....)))))))..)))))))))....))))..))).)))))...))))))))))))))))))...))))))).)))))
.....))))).)).)).))).)))))))).........................((((((....))))))((((.(((((......(((((((.((((((........(((((((((..................((((((((.((((((.((..((((.(((......)))))))..)).))))))))))))))....(
((((.((((......))))..))))).(((((((.(((((((......)))))))(((((......))))).(((........))).((((((....))))))...)))))))((((..........)))).................((((........))))....)))))))))))))))..)))))....))....
.))))).)))))))))))))..))))))....))).)))))))))))))((((((((..((....))..))))))))))))))))....))))).........(((...((((((.((((....))))..(((((((((.............................((((((..((((((..(((((((....(((((
(..(((((..((...(((((.((((...)))).))))).(((((......)))))...(((((.(((......))))))))..))..)))))..))))))(((........)))..(((((((((.(((..(((((............)))))....((((((......(((((.((((((((...(((((((...))))
)))........(((((..(((((......(((((((..((((((((((.....((.(((.....))).)).....)))....................(((((((.............))))))).(((((((((((...(((((((((((((..((((((((((((((..(((..........((((((........))
)))))))))))))))))))))((((.........)))).))))))).))))))..(((((..((((....((((((((....(((.((((...))))..)))....))))))))....)))).))))).(((....((((.......))))....)))............)))))))))))......((((((((...))
)))))))))))))..)))))))(((((((((((...........)))))))))))((((((...(((((..(((((...)))))..))))))))))).....((.(((((((((..((((((..........((((((((((..(((.((((...((((....(((....(((.......))).....)))...)))).)
))))))..)))))))))).........))).)))..))))))))).)).......................(((((((((((.......((((....))))........))))))))).))...((((((((...)))))))).....(((.((((.....)))))))...)))))..)))))....(((((..((((.(
(((.((.....((((((...((((..((((......))))..))))....)))))))))))).))))..))))).....)))))))))))))...))))))...(((.((((((.....)))))).))).........)))..)))))))))......................)))))))..)))))).))))))....
.....(((((((((...........)))))))))....))))))))).........)))))).))).....
You can download the minimum free energy (MFE) structure in
II. Results for thermodynamic ensemble prediction
1. The free energy of the thermodynamic ensemble is [-595.38] kcal/mol.
2. The frequency of mfe structure in ensemble 4.49126e-37.
3. The ensemble diversity 683.77.
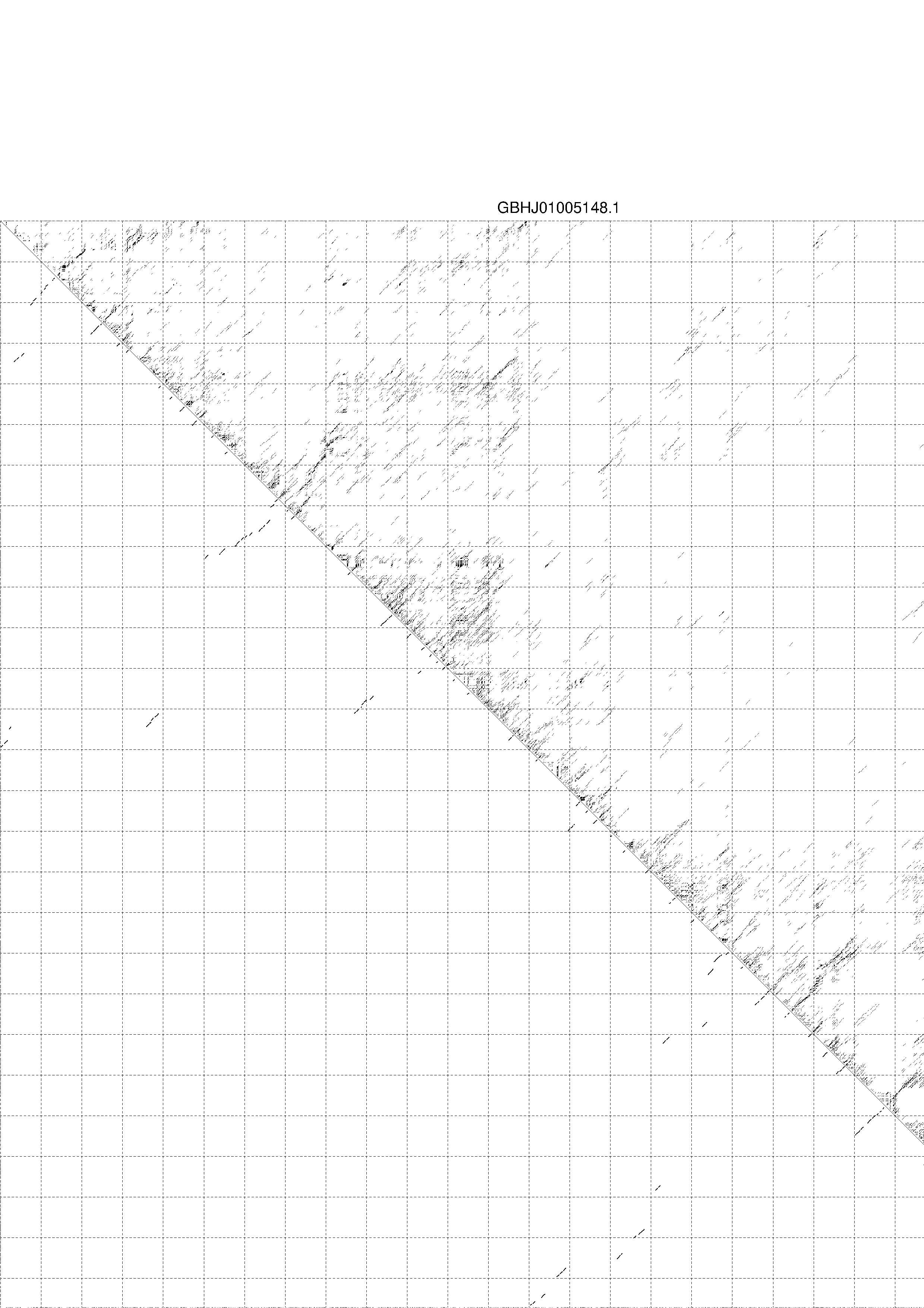
4. You may look at the dot plot containing the base pair probabilities [below]
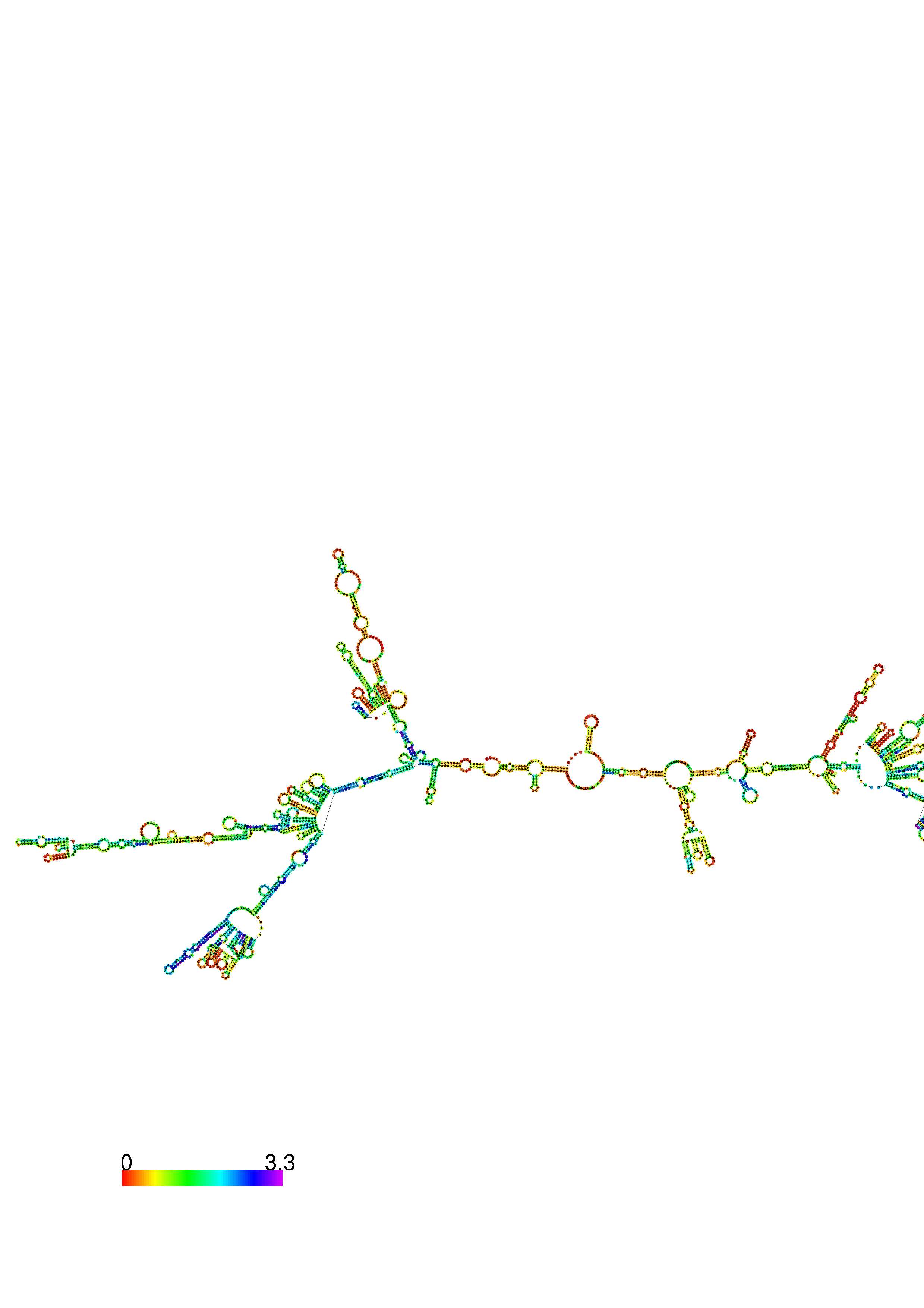
III. Graphical output
1. A note on base-pairing probabilities:--
The structure below is colored by base-pairing probabilities. For unpaired regions the color denotes the probability of being unpaired. The reliability information of RNA secondary structure
plot in the form of color annotation, Positional entropy ranging from red (low entropy, well-defined) via green to blue and
violet (high entropy, ill-defined).
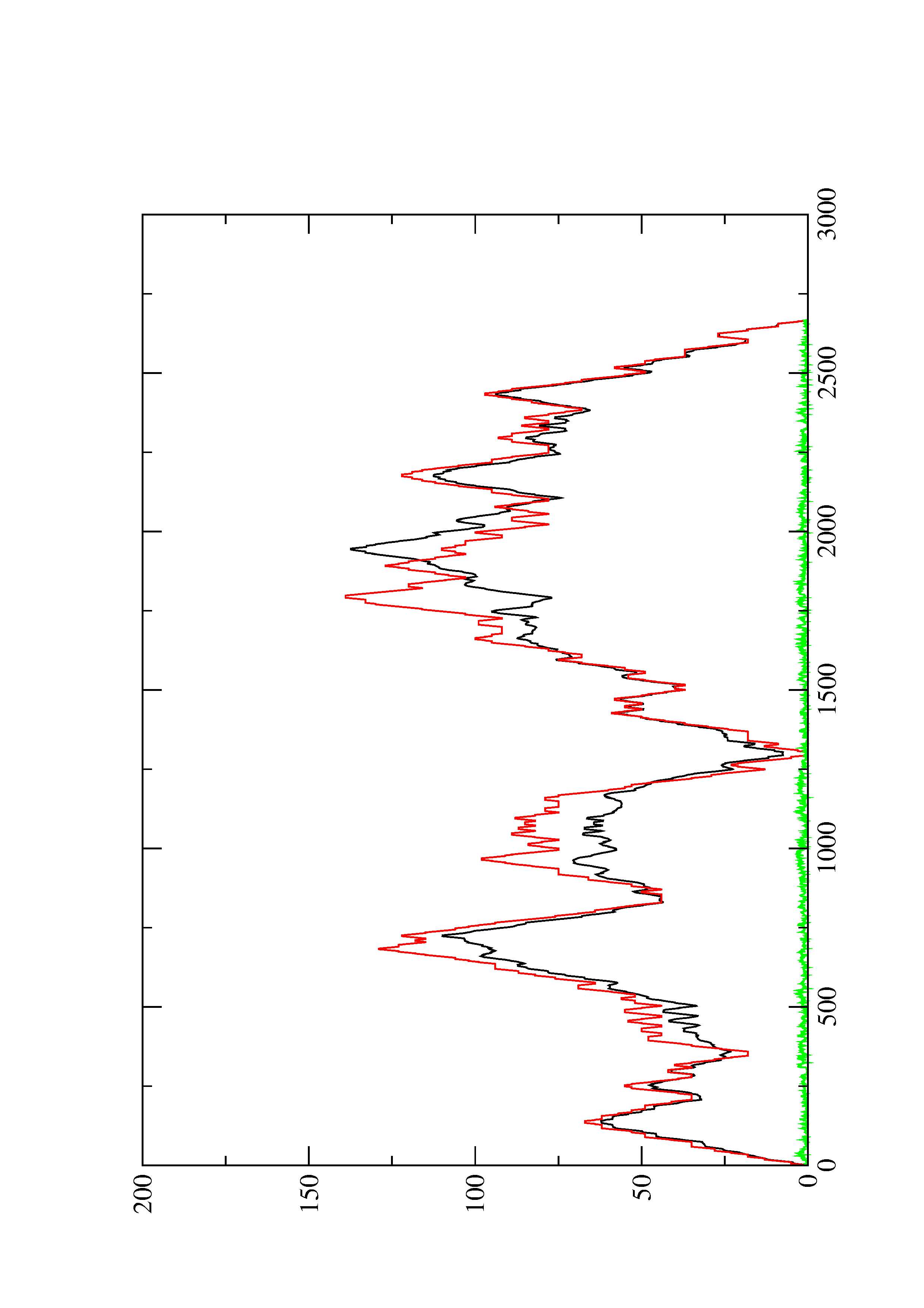
2. Here you find a mountain plot representation of the MFE structure, the thermodynamic ensemble of RNA structures, and the centroid structure. Additionally we present the positional entropy for each position. The resulting plot shows three curves, two mountain plots derived from the MFE structure (red) and the pairing probabilities (black) and a positional entropy curve (green). Well-defined regions are identified by low entropy.