lncRNA Structure Prediction
Species Name : Withania somnifera Sequence Id : GBHJ01005498.1
I. Result for minimum free energy prediction
The optimal secondary structure in dot-bracket notation with a minimum free energy of (-396.40) is given below.
1. Sequence-
GUUAUGAACUUGAUCGAAUAGGGUUGUCACAUAAAUAGUAGGUUAGUCAAAGUCUAGCAG
AAGAGUUCUCCUCUACAUUGUGAUUGUCUUUUCUCUACAAUUAUAAAUAGAAAUCCUCUA
CAAGGACAAAGAACACAUCAAAAGCUCAAGAAAAGCCAGUAAAAACUCUCAAGAACAAAA
CAUUUCUAGUCUUCCUCGGAGUCCUGGAGUUCCAAGUCCAACAAAGAAGUUCCAAAUCAA
GAUUUCAGAACGCGACUAGUUCUGCUGCGGGAUAAAGUGAUUGGAACUCAAACUAGGUGA
CCAAUACCAAACACAGCAUUCUUCAAUCCUCCAUUCACACUGAACAUCAAUGGCGAUGAU
UUCAAUUCAUGUUCUUCAAUUUGAGGACAAUGAUUUAACUGCUUGCAUCAGGAACUUCAU
GAGCCGCUCCAAUUGUGCAAUUAGUCACAAAUUUUACUUGUACCAUGCAGCAGAGUAAUU
UCAACCUGAGAGAAGAAAAUGCAAGUAGUUAGUUGUAUGUGUUUCAUUAAGCAGAUAUCU
AACCAAAUAACUAGAUGCAGCAAGAAAGUUUUGACCUAUCUGACUGGAUUCAAACUUUCG
AUACCUUUUAAACUCCAUUUAGUUCAGUAGAAAGAAAAUCAGAAAAGAUGAAGAAUUGGA
AGAUUUCACUCAAAUAGGUAAGGAGUUUUUGGCCAAAAACAUCCCUAAAGUAUACUAGUA
ACUUCAACUACAUCCUUAAAUAUCCCUAUAAGCAAAACAACAUCCCUCAAGUAUAAUCAU
AUCUUAUCACUGUAAGCAACAAAACACCCUUCAAGUUCAUUGAAAUGAGCAGGUUACAUC
CUUCUGUUAUUAAUGGUUACAAAAUUAACAAAAUCCUCAAAAUCAUGUGCAAGACAUGUC
AUCUUCGCCAAAAACACCAUGGUUCCCUCAAAGAUUUUUGAAGUCGGUACAUAACUUGAA
CCACUUAAAUCGUGCUUUAAACAUGAUUGGAGGAGAUUUCAAGGGAUAUAUCCCUUUCCU
AUUGUUGUAUAAUUUGUUGUGAAGAAGCCAGCCAAUUCAUCCUUUUCAUCACUAGUAACA
UGAACAGUCAACAACAUGCAACAGAAUUGCAGAGCAUAGUAAGAAAAAGAUUCCCAUCUC
UUCCUCUGUCUCACCAUCAAGUCAUUAAAGAAAAAAAGAAUAAAACCCACUUCCUCCAUU
CACCAUUAUCCUUCACAAGAAAAAGAUUCACAGUAUAUAUUUGUAAGUUCUGUAACAAGC
UUUCACUACAUUAGAUCCUAAAAAGUGAACUAAUAAUCCAAACGUAGAUGCAGCAUCCAA
GAGUGUGGGCCUAGUGGUCAAUGAAGUGGGUUGAGCACCAUGAUGUCUCAGGUUCAACUC
CCAGUAGAGAGAAAACACUAAGUGAUUUCUUACCAUUUAUUCUAGCUUUAGCGGACAAAG
UUAUCUGGUAUCUAUUGGUGGAACGUAUCCCGUCGAAUUGGUCAAGAUGUAUGCAAGCUG
GCCUCGACACCACAAAUAUCAAAAGAAAAAAAAAAAGAACAAGAAAUAUAGGAACUCCAG
AAAAGCAGAAAUAGAGCAGCCCGGUAUACUAAGCAUCCUACUUUCACGCAGAUUGUAUGA
ACUUCUUCUCAUUCACUGUCAUUCCCACUCUUUGCUCCAUUAGGUUCGAUUUUCAACAAA
UGAAAAAACACCACAGUCGAGACUAUUAUUCCAAUUUAGGAGUGCAUGAGGAACACGUAA
CCCUAUUGCAGGAAGAAAAUCUCUGUCUCAAGUCUUUUCCCUAUAAUGUUUGGAUUUUGA
AGGAGAAUAGAGGUGGUAUUAGUUUAACAGAUUGGUGAAUUGAAAUAUCUAAAUAUGUAU
UCUAAAAAUUAGAGGACUUUAUUAUCAGUUGAAGUCAAUUCAAAACGUUUAUCUAACGAU
AACACUAGCUAACAUUCCUUCACUCCAAAAAGAGAAAAAAAAAACUAACUACAUACUAAG
GUUAACUUUUUAAACAAAAAAAGAAUAAACGGUAAAUAGGACGCACACUGUAAUUUAAAA
AAAAA
2. MFE structure-
.......((((((...((((((((((...............(((((.......)))))...((((((....((((..((((((((((.........)))))))))).))))..((((.....)))).................(((.......))).......))))))..((((........)))).((.(((((((((
((.((((((((((..............((((((((.(((..((((((((((.......)))))).....))))....))))))))))).......((((.....))))....((((((........((((((.((((...))))......((((((.(((..((..((.(((.......((((((((........(((((
((((((((((((.............(((.((...((((((.(((.........))).))))))..)).))).............)))))..........))))))))))))((((....(((.((((((((((((.((.(((((.....(((((((((..(((((...)))))((((((.(((.((((......((((((
((..((((..............(((........))).............)))).))))))))..)))).))).))))))..(((((((((....)))))).))).....)))).)))))...............................................................(((((....)))))....
..............((((((....)))))).))))).))...)))))..)))))))..)))....))))....)))))))).......))).))..))..))).))))))..(((......)))...(((((....)))))(((.(((.((.....)).))))))..(((((((.......)))))))))))))......
((((((....))))))......))))))...((((((.(((..((.((((((..((.(((((((((..((..((((.((.(((.............(((((.....)))))(((.((((...(((..((((....)))).)))..)))))))....))).)).))))..))..))))).....((((((((((....(((
.(((((((.(((.(((..(((.....)))((((((((....))))....)))).....(((..(((((((((((.((((....)).)).))))).........)))).)).)))........))).)))..))))))).)))))))))))))((.((...(((((......((((.....((((((((((((((.(((..
.))).))))))((((.......(((((((.......)))))))..)))).))))))))..))))......)))))...)).))..)))).))....)))))).))..))).)))))).................................)))))))))).......(((((((....((..((....))..))...)))
.))))..((((..((.((((........)))).))))))..............)))))..(((..((((((((((.....)))))..........)))))..))).((((((......))))))...)))))).)).))))))))))((((((......)).)))).))))))......(((((....((((((...(((
(((((..(((.((((.((((.(((....((((...((((((((.....((.((((.((.((((((...)))))).))..))))...))......))))))))....))))....))))))).))))..)))...)))))))).))))))................(((((.........))))).((((((......)))
)))............))))).........................
You can download the minimum free energy (MFE) structure in
II. Results for thermodynamic ensemble prediction
1. The free energy of the thermodynamic ensemble is [-435.98] kcal/mol.
2. The frequency of mfe structure in ensemble 1.28937e-28.
3. The ensemble diversity 581.50.
4. You may look at the dot plot containing the base pair probabilities [below]
III. Graphical output
1. A note on base-pairing probabilities:--
The structure below is colored by base-pairing probabilities. For unpaired regions the color denotes the probability of being unpaired. The reliability information of RNA secondary structure
plot in the form of color annotation, Positional entropy ranging from red (low entropy, well-defined) via green to blue and
violet (high entropy, ill-defined).
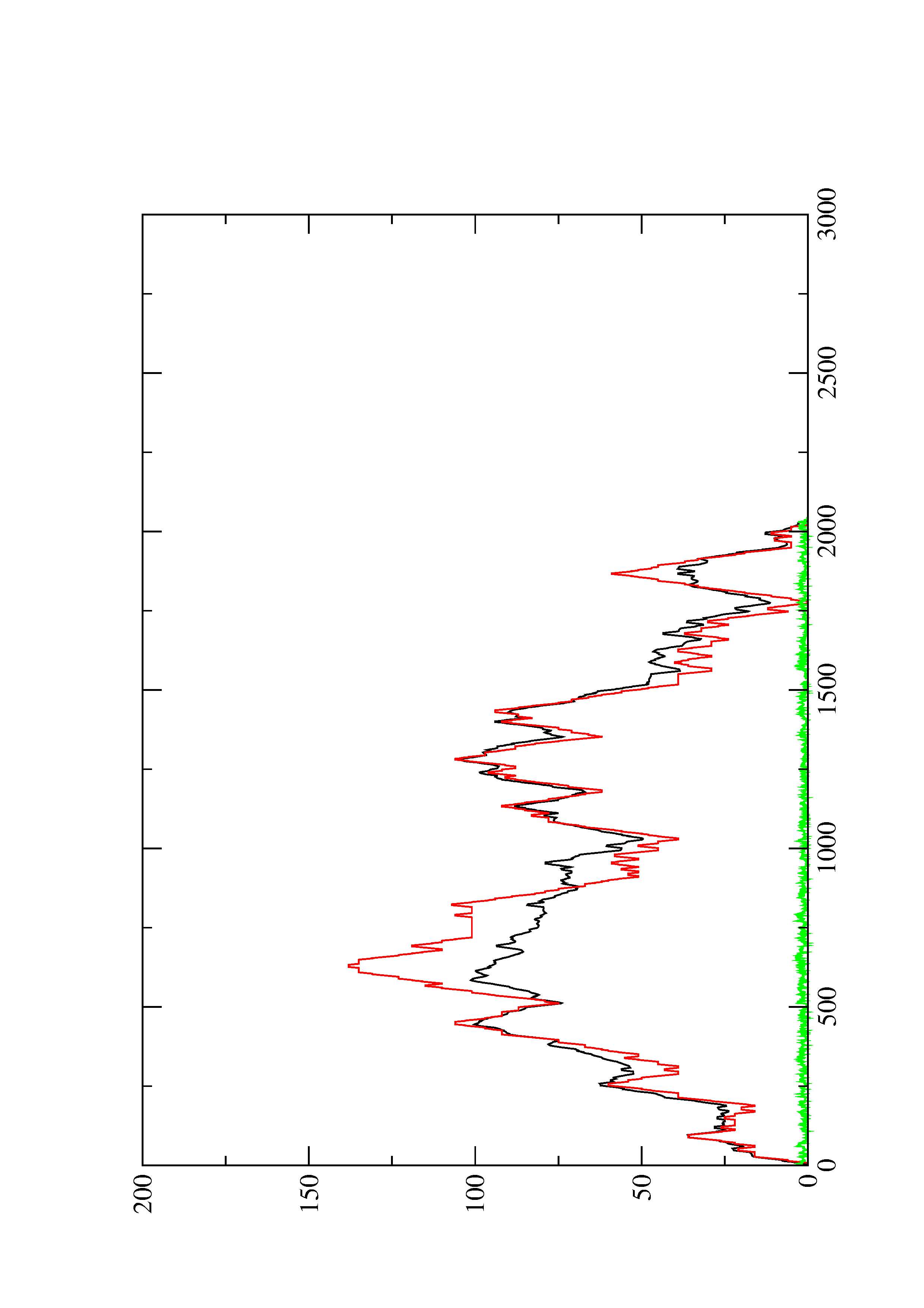
2. Here you find a mountain plot representation of the MFE structure, the thermodynamic ensemble of RNA structures, and the centroid structure. Additionally we present the positional entropy for each position. The resulting plot shows three curves, two mountain plots derived from the MFE structure (red) and the pairing probabilities (black) and a positional entropy curve (green). Well-defined regions are identified by low entropy.