lncRNA Structure Prediction
Species Name : Withania somnifera Sequence Id : GBHJ01006353.1
I. Result for minimum free energy prediction
The optimal secondary structure in dot-bracket notation with a minimum free energy of (-505.71) is given below.
1. Sequence-
GUUAAAUUGUCAAUUUAUCAAACCACUAGCCAGUAUGCAUCUAACAUGUAUCACAAGGCA
ACAUACAUCUCAAGUCCACCAAAAGUGAUGAGUUCGAUAGUAAUUUUAUUUGAAAUUCAG
UUCCAUUUAGGAACUGAUUCAGAAAGGCAUGAGGAUUAGAAAGAACAGUUGAUGCUGUUG
CGAAGAAAUUGUUUGUUCCGUCUUCUUAUUUCUUAUCUCAUCCCAAAUUCUUCUGUCAAU
CCUUCUAAUUUCAGCAUCUCCAAACACAUCAUCAUUGAUAUACCAAGAACAUCAUAGCCC
AAGGCCAACUUGAAAUAACUCAAAUCAACAAGAACCUACUUUAUGAACUCUUGCUUUUUU
UUAGAGCACUGAGCCAGCAUAAGCAUUACCUAAAAUAUUCAGAACCAAAAUCAUAUUUGC
UCAGAGAGACAAGGCUGUGAUAAUCAGAUAUACUCAAUCAUCAAGCAACUUUAUCCUGAA
CUAUAUAGGAGAAUCUUUAUAUCGCCUUUGAGCAGCCUUCAAUAACAUUGCACAUUUGAU
GGUAAGUCAAAACACUAACAAUCUAACGUAUAACAACCACCUCAAGAACCUAAAGUUAAG
ACUCUAGUCAAUACCACAAAGCAAGGUCUAGGGCUUACUGCAGAGAUACAGUUGUCAUCU
CAAAUAGACCCUAGAUGUAAACUGCAUGCUUUUGGAGAAUUUGGAGACAAUCACCCAUUG
ACAUUCUUGAAAUUGGCCCUUGACUCAAGGAAGAAUAAGCCAAAGCAUCCAUAUCAUUGU
CCACAAAAGGACAAAAAUGUAGUGCUGUUAUUUAACUCACAAAAUGUAGGUACAAAAGAA
GAAGCUGAUGACAGCACUUACACUAUAUGAGUUUAUCAGACCCUACACCCAAGUUUAAAG
GAGUAAGGGUAUAGUGAACUCAAUUGUUUCAUCAUCAAGAAUUCAAACAAAAAUCUCAAU
UGUCAAGAUUUAGAACAAUCAUUAACGCCUCAACCGAAUCCAAUAUGAUCAUUUGUAAUG
UAGCUUCACCAAGAGGUUCCCCCUGCUUAUGCAGGGACUUCGAUCCACUACACCUCAUAU
UACAUGGAUUGAAUCAUCAUCCGCAAACCAGACAACAGAGAAACCAAGCAAUCUUUACAA
GUAAUUUAGAAAUUGAAUAACAGAUAAAUGUAGCUGCUCUAGUGUACUCCAAAACAUAAU
GAGAAAAGGGGUUAUGAAGGAGAACUAUAAAAUAGAGCAACACACUUACUAUUGCUUCAA
GGGAACGAGUUUAUCAAAAAUGGAUUAUUUCUUGACAACCAGAUAAAGAAGCCUUCAAAU
CACAACUUUUACACGAUUAACCAGGUAAAAACAACAAAAAAGAUAGCAACUUGAGAUUAU
UCAUAUCCAAUUUUAACUAUAUAAUACUUCUUCCAUUAUAAAAAAGAAAAGCAGCCCUAU
GCACUAAACUCCGCUAUGUGAGCUCACAACCAACACUAAACCCUUGUUAUCCAGCCGAUU
CAUAUCAAAUUCUAAUUGUAUAAUACAAUUUCACCAAAAAAAUUGGCAAUCCAAUGCACU
ACGCUAACGCUAUGUGCAGGGCCUAAGAAGGGGCACAUAGCCAAAUAGGACUCCAAAAAG
GCAACAAUAAACCCUUGUGAUCUGGCUUAUUCAUAUCAAAUUCUAAUUGUAUAACAACCU
CAUUAAAAAAAUAGCCCGAUUGCAGGUCUGGGCACAACCACAAAGAACUCAAAAUAAACA
ACAAUAGGCCCUUGUGAUCUAACACUCAUCAUUUGUAAAAUUGGAACAAGGAGAAAUUAA
AAAGGCCUCAGAACCUAAAGGUAAAUGCAACAUCAAUGGAGCUUAAUAAAGCCCCUCAUA
UGGUACGGACAAUUACCAUGCAUCACCGGCCACAAAUAAACAUAAGUCAUUAUCCUUAAC
AAACACUACUUUCAACAAUUAACACAAAAAUCUAAGUUGCUCAAAACUCUCCAAAAGUGC
ACUGUUUUUAGCAUAGGUAAAGACAACUAGUUUAGCGAAGUAGACAAAACAUUUUCUAGU
AAAACAAAAGCUAACUGUAAAAUAAAGUAUUCUAGGCUCAGACAAACGAACAGCUCUAUC
AAUCACGGAAUAUCAGAAGCAGAACUGGCAUUAAAGCUGAAUUUAUAUCAUCACAGCCAA
CAUUCAAACAAAAUACAACAACAUACUCAGUAAUUUGCCACCAAGUAAGGGUAAGGUAUA
CUCAGUCCAUAACACGAACCCAUAUGUGAGGUAGCGACGCUGACGCUCCGUUCAUGACAC
ACCAUAGUCUAGUAAUAACAAUAAACAAUAUAUAGUUUUCGAAAUAACAUGCAAAUAACA
AAAAUAGCAUCAGAGAUAGAAAAUAACACAAUAAUAAAAAAUAGUACCCUAGUACGAGAA
ACGCCCUAUCAGCAGAAAGUCGCACCUACUCCUAACCUACCACCCUAAUUCGCCCUACCA
CACCCUCUCGCACCAGCAGACUGAAGCAUCCGAGCCCCUCUUCGUCAUGCCCAAAUCAUC
UCACACCAGCACACUGAAGCAUCUCAACCGUACUUCUCGCAUCCACCCUGUCACGAAUAA
CCACAUUAAUACCUACGCCAUUGAAUAAAGAUUAUUGAGACGCCACGGAAGAUGAGGUGG
UAGGGUUUCUACCAAGAUUAAGAAGUGAUUUUGAAGGCCCGGG
2. MFE structure-
.....(((((((((((((((........(((.((((((((.....)))))).))..))).....((.......))..........))))))))).)))))).......((((((..((((((((.....))))))))))))))..(((..((((((..(((.((((((((..(((....)))....)))))))).))).)
)))))(((((((((((((...........((((((....(((.........................((((....))))....(((((...((((((....(((....((((................))))..)))...))))))..)))))........((((((.(((((((.((....))................
...............((((((.(((((...((((((((..(((...((((((.....................(((((.......))))).......))))))....))).))))))))........((((...((((.(((((((((..(((........(((....................))).......)))..)
)))........))))))))))))).((((((((((...((.(((((((....)))).)))))...)).))))))))......(((..(((((((((((((((((.............((((.((((((((..((((((((((...)))))........))))).............((((((......))))))...(((
((((((((((((....((......(((....)))......))....))))))))))).)))).....(((((((((...(((((((.((..........)).)).)))))...)))))))))............)))))))))))).......((((..(((((...(((..(((..(((((.......((....))...
....)))))..)))..)))..((((((.....))))))..((((((....))))))..(((((((((.................))))))))).................)))))..)))).....((((((.......(((((...............(((......)))...(((((((..((.((((....((((((
..........))))))..)))).)).......)))))))......)))))))))))((((((((...(((((((.....))))))).))))))))...))))))......((((...((((.............))))....))))..................((((((((.(((((......................
.......(((((...........)))))..(((......)))..............((((....)))).............((((((..(((((((((((.............(((((((...)))))))..........)))))))........((((...((....))...))))((((((((..(((((.......(
((.....((...)).....))).........)))))((((((.((((.............................................)))).)))))).(((.((....))))).........................)))))))).(((......)))................)))))))))).........
...((((............(((..((((.........((.((((....)))).))....((((((........)))))))))).)))))))...........((((........))))..............................)))))))))))))......)))))))))))))).((((((((.......)))
)))))....(((((......))))).......)))))))))))..........(((.........)))......)))))))...........))))))........)))....))))))......(((((.....(((((..............................................)))))....)))))
.......(((((.((((...............((((........))))((((.((((((((((.....((((...(((........))).........................((((((........((((..............))))........))))))...................((((....))))....)
))).....))))).....))))))))).......))))...))))).....((((((((((....(((.((....))))).....((((.((((......))))..))))................(((....)))...(((((...((((...(((((.(((.......(((...........................
.))).....)))...)))))....)))).)))))..))))))))))........))).))))))))))........)))....
You can download the minimum free energy (MFE) structure in
II. Results for thermodynamic ensemble prediction
1. The free energy of the thermodynamic ensemble is [-550.86] kcal/mol.
2. The frequency of mfe structure in ensemble 1.52869e-32.
3. The ensemble diversity 618.63.
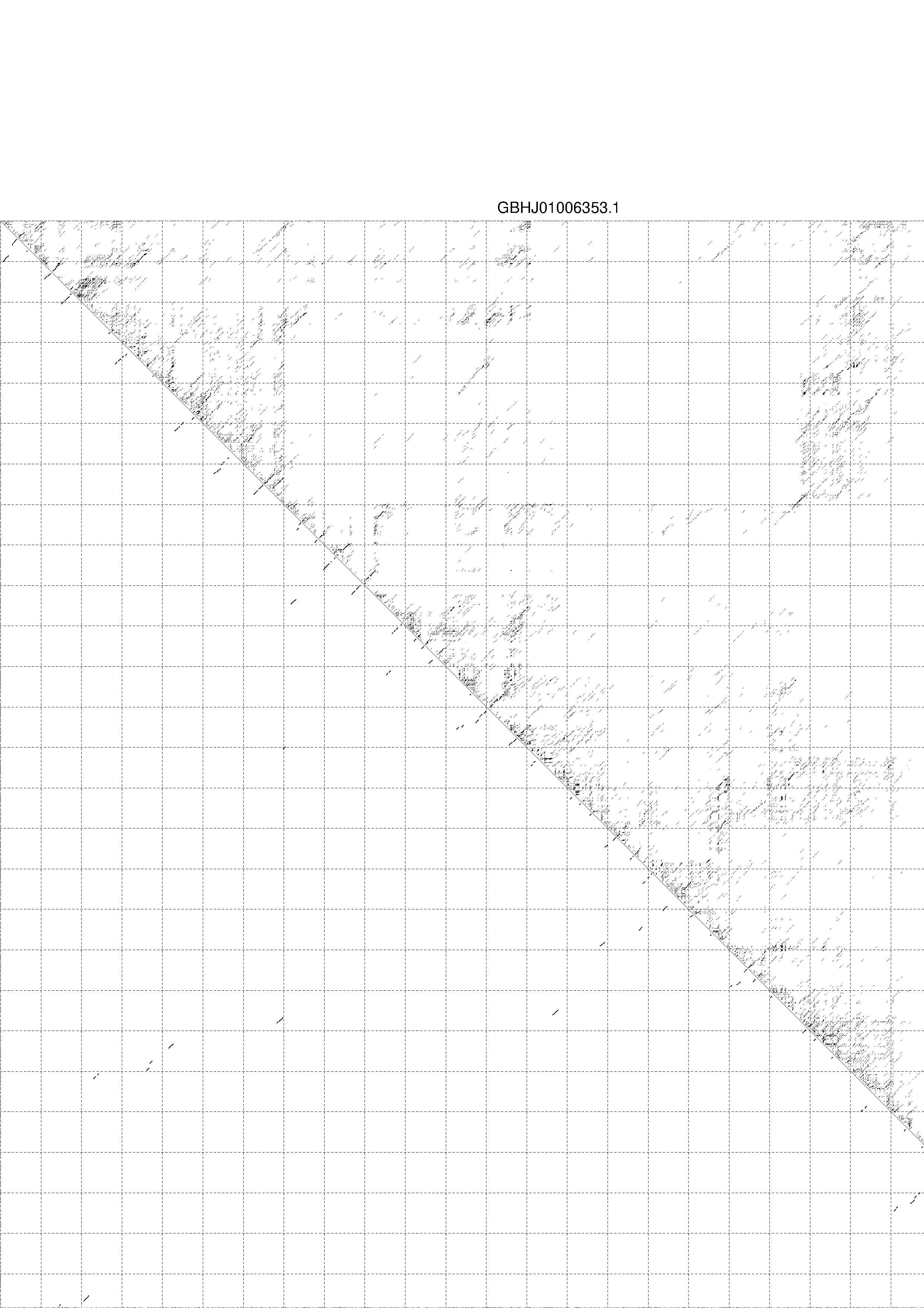
4. You may look at the dot plot containing the base pair probabilities [below]
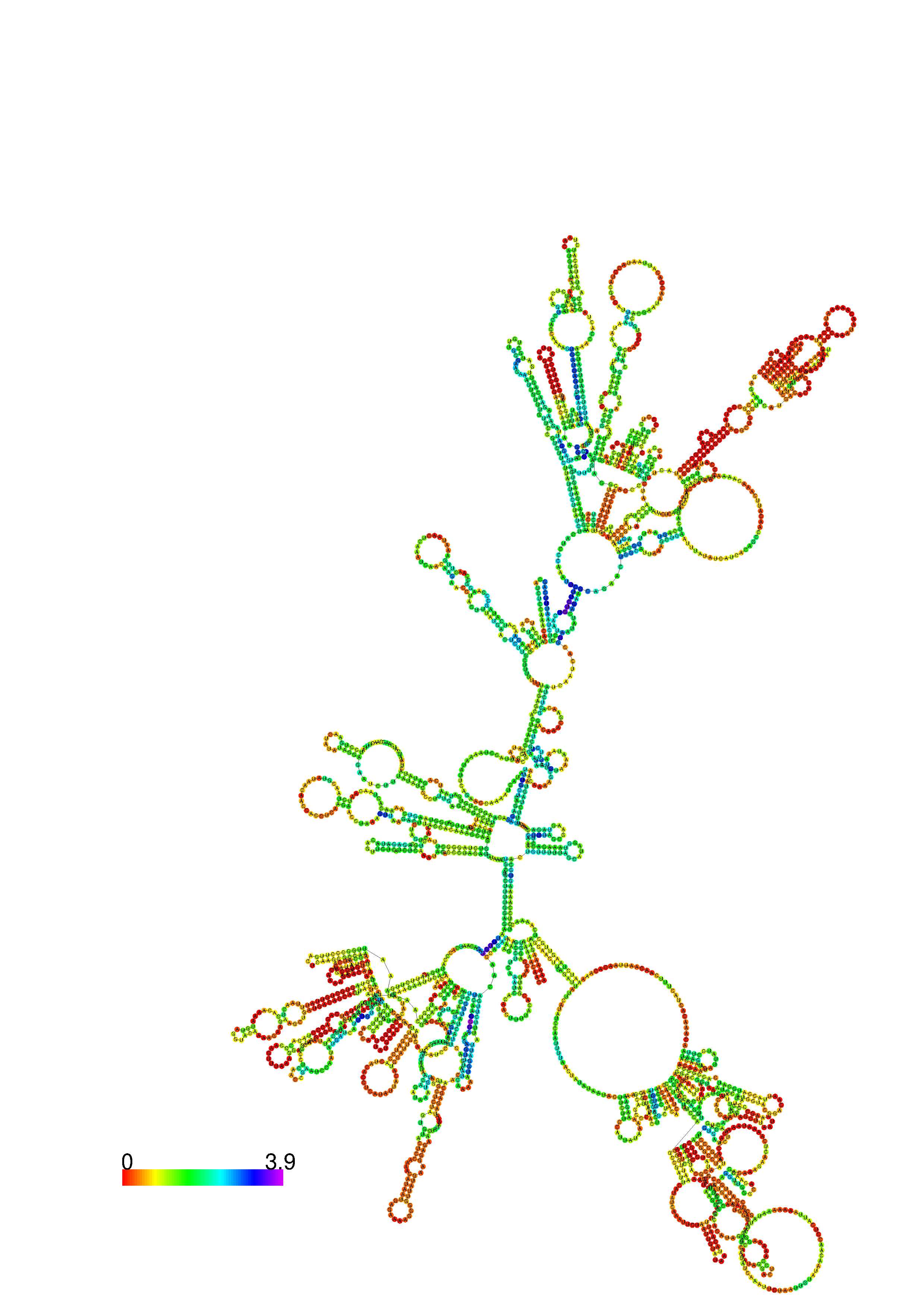
III. Graphical output
1. A note on base-pairing probabilities:--
The structure below is colored by base-pairing probabilities. For unpaired regions the color denotes the probability of being unpaired. The reliability information of RNA secondary structure
plot in the form of color annotation, Positional entropy ranging from red (low entropy, well-defined) via green to blue and
violet (high entropy, ill-defined).
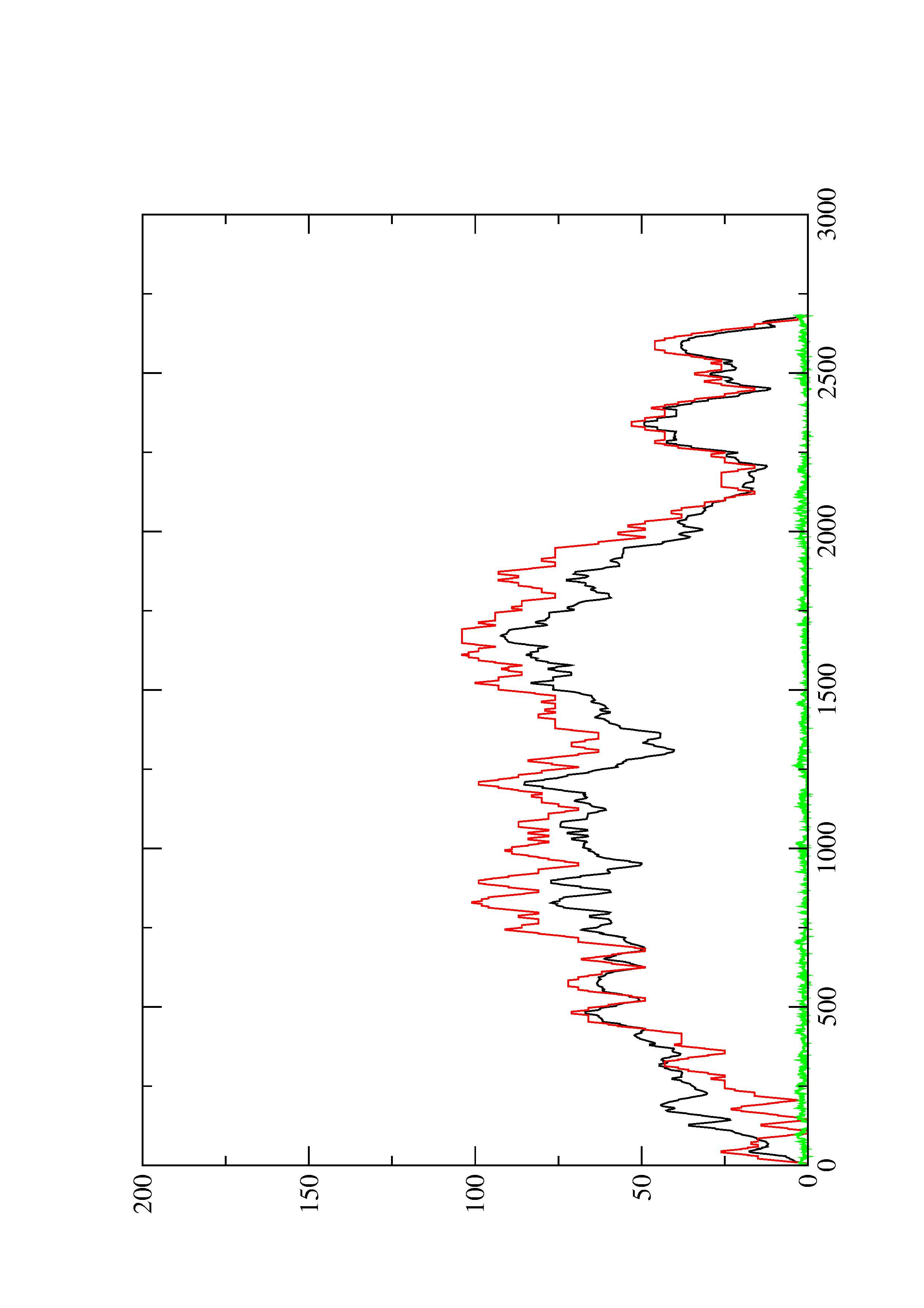
2. Here you find a mountain plot representation of the MFE structure, the thermodynamic ensemble of RNA structures, and the centroid structure. Additionally we present the positional entropy for each position. The resulting plot shows three curves, two mountain plots derived from the MFE structure (red) and the pairing probabilities (black) and a positional entropy curve (green). Well-defined regions are identified by low entropy.