lncRNA Structure Prediction
Species Name : Withania somnifera Sequence Id : GBHJ01006695.1
I. Result for minimum free energy prediction
The optimal secondary structure in dot-bracket notation with a minimum free energy of (-517.60) is given below.
1. Sequence-
AGAGAACUUAAGUAUUCUAGUGAAUGAUAAUGCUGGUAAGUCUCCUCCUUAUUCAGCCAA
UCUUAUUAUCGAUUUUUUGUUGAAUCUUCUUAAGUUUGUAAUUCAACUACAUCUUUGUCC
UUCCAUCUUUCAAUCUUUUGCAUGCUUUGGACUAGGAGCUAAGUGGCAAAUGAAAUAUUA
ACAAUACUUUUGAUAUUCUUUCUACUUUCUCUGCAAACUAUUCCAUUUGAUUGUUCAAAG
GCAACUUCUACUAAUCGGAAUGUAGGUUGAGUGUAGUAAGUUUGACUUCCUCUAAUCAAA
UGUUUCAUGAGUCGAAGGCUUGAAACUUGAUGGGUAGUGGAUCCCCCUCUACCCUUCUCC
ACUUAAAUACCAUGACUUUAUCUACAUCAAUACUCGAGCAUUGUGGCAUGCGCCUAACCC
ACACAUCUCACAUUUUCCUCUUACCAUUAGAUCAAAACCCUGGGGGCCUAGUUUGUCUAA
AAUUUUAGUUUUACAAAUGAGUUUCAUAAGGUAAAACAAUAAUGUGCUCCAAAAUCAGGG
AUGCAGGUUUGCCUUCUGUAGUUCUCAAGAUAACUUGAUUUUUAAACACCCUUCAGGUUU
GAUCUAGUUGCUAAAUUUCAUUUCAUCACUUCCCUCCCUCUUCCAUUCUCUGAUCAUAAU
CAACUCUUAAUAUAAUUAUCUUCUACAGCUGCUCAUCAAUAGCAAGGAACAUUGGAACCG
UGGCAAAUAUUUUCACCAGUUGUAUGUAUUAGCUCUAUUAGUAUAUUAGAUAUACAAAUG
AGUUGCAUAAGGUAAAACAAUAAUGGCUAACUCCUUCUCAUGUUUGACGUUAUAUGUUGU
AUCCUAUAUACUCAUAGAGCUAAUCCAGAUUUCUGUUCCAUGCAGUGGACUCUGAUGAUG
AUGGUGUUGCAGUAAUGCCUAACCUGAUCUAGUUAAAUAUUAUAUGCAACAUAAUGUUCA
AUUACUUUUAUCGUAGUUAUUUGACUUGAAUAUAUCUUUUUGUAGUACGCUGGAGAAGAU
GAUGAGAGCAGUGAUAAUGGAGGAAUGCUCUGUGAGCUGUAUCCUAUCUGAAUGCACUAG
UUUACAUAUAGCUUUAAUUUGUUGUACACAUCAGAUACAGUGUUAAAGUUUUUCAGUCAA
CUCUUCUUACAAUUCUGUCAGUGUGCUGCUGCUCUCAAAUACUUAUCCAGACCAACUUCU
UGCUUCUCAUUUUGUUGCUUAUAUGAACAACUUUUUAUACUCGACUGACAGUGAUGAAAC
AAGUAACUUAGAGAAAUUGGAAAAAUGCGAGUGAAAGGAGAGGAAUGAGUAUCUACAGGA
GGAUAAUGCUCGGAGUUUGGAAAAACUUUUAAGGAGGAGAAAAUGAGAAAAUAUGAGGGA
CAAGGAUAAGUUACUACACUUUCGUUAAUUCCUACAGGGAAACCUCUCCUCACUUGGUUA
AAACUUUUUGUAAUGCUAAAACACACCUUAGAAGCGAGAAUAAGAUGGACAUACGAUAAA
UAGAUGAAAUCACAGAACGCCCGUUAGCUUUAUCUUGGUCUAUUGGCCCUCUAUAUCUCU
UUUCUUCUCUGCCGGUGUCUUUAUUCCUUCUCCUUGAUCCACCAGUAUCUGCCUGACAUU
UCCUUUCUCUCUUCUAAGUUACCAAAACAACAUUUAUAUAACUGUUGAAUUUAUUGUGUA
UGCUAAUUGUUUACUUGUGUAAGCGCGAAUAAAAGUUCCAUUUUAUUUAUUAGGGGAUGG
AGUUUUCCCAGUUGUGACAUUAAUUUUGUCUCUGAAUUGCAGAUCUUCCAGACCUGGAAA
AGGCUAGAGGAAAUUAACGCCAGCUGCUGAAAACUUUGGAAAGGGGCUGAUGAUCAGCAG
AGGUCUUUAGAGUUUAGUCUUUUAGGGAACAAAAUAUAAAGACUUUCUUGCUGAAGUAUU
CAGAUGUAUCGAGGUAGUCGGAAGGCUGAUUUUAGGUCAUGCAUUUGAUCUAGUUUCACC
UUGUCAGAUUGGGUACCAAGUCCCGUUUGUUAGCUAGCACUGUAACCCCAAUAUGAGACU
UUCCAAAAUAAUUUCGGAUUUAGUCAGGCUCCCAUGUGGUUCCGAACAUUGGGUGGAAAA
CCAAAAACAAAAAAAGAAAGUUUACUUGGCUUUUGUAAUUCUUUGUUUUCUUUUC
2. MFE structure-
(((((((..((((((((....))))).....(((((((((.......)))).)))))(((.((((.....(((((......)))))....)))).)))...........((((((((((..((((((((....((((((((...((((((((..((((..((((((.(((.(((.((((((........)))))))))))
))))))).))))....................))))))))))))((((((......(((((.(((...(((((((((((.((...(((((((..(((((..((((((..((((...)))))))))))))))((((((.((.....)).))))))((((((.(((((............................((((((
(((((((....)))......................((((((...(((((((((((..((((((((....(((((...((((((..((((.......((((...((.(((((((.......((.((((((........))).))).))))))))).)).....))))....)))).)))))))))))..)))))))))))
))))))))........((((((((.((..(((.(((....(((((((((((((.....................((((((((((.(((((((((((((.((.((.((..(((((....(((((.((..........(((..((.((.(((((((((((.(((((((..((((((((.(((((((((((((((.....((.
...)).......))))...))))..))).)))).)).))))))..))))))).))))))))))).)).))..)))))))))))))))..)).)).)).)))))((((((((((((((..((((........))))..)))))).)))))))).((((((((((((.......))))).........))))))).......
...)))).)))).))))))))))(((((((((....((((((.....))))))(((((((((.....(((.......))).....))))))))).........((((((..((((....(((....((((........)))).....)))...))))...)))))).)))))))))......................((
((.((((((((..((((..((((.(((....))).))))..))))....))))).))).))))....))))))))..)))))......))).)))..))..)))))))).......))))))))))))))))((((((....)))))))))))..)))))).............)))))...))...)).))))))))))
)...))))))))....((((......))))...(((((....((.....))...))))).........)))))).))))..)))))))).....)))))..))))).....((((((.(((.((.(((((..((((((((....)))).....................(((.((((((................))..)
))).)))...(((((((...(((..(((.((((((((((((((((..(((((..........)))))......))).))).)))...(((((((....))))))).((((((((......))))))))..))))))).)))((((((.((((((.((((.......))))((((.....))))(((((.((.(((.....
))))).))))).........))))))..))))))(((((((((...((((...((((((((((((((((...(((..((((...))))..)))...))))))))))..)))))).....))))......(((.....))).((((.(((..((((((((......))))))))..))))))).(((.((((((.....((
(...(((....)))....))).....)))))).)))..)))))))))........)))....))))))).(((((((........))).))))(((....)))..........))))))))).))..)))))))))....))).)))))))....
You can download the minimum free energy (MFE) structure in
II. Results for thermodynamic ensemble prediction
1. The free energy of the thermodynamic ensemble is [-561.92] kcal/mol.
2. The frequency of mfe structure in ensemble 5.86543e-32.
3. The ensemble diversity 483.77.
4. You may look at the dot plot containing the base pair probabilities [below]
III. Graphical output
1. A note on base-pairing probabilities:--
The structure below is colored by base-pairing probabilities. For unpaired regions the color denotes the probability of being unpaired. The reliability information of RNA secondary structure
plot in the form of color annotation, Positional entropy ranging from red (low entropy, well-defined) via green to blue and
violet (high entropy, ill-defined).
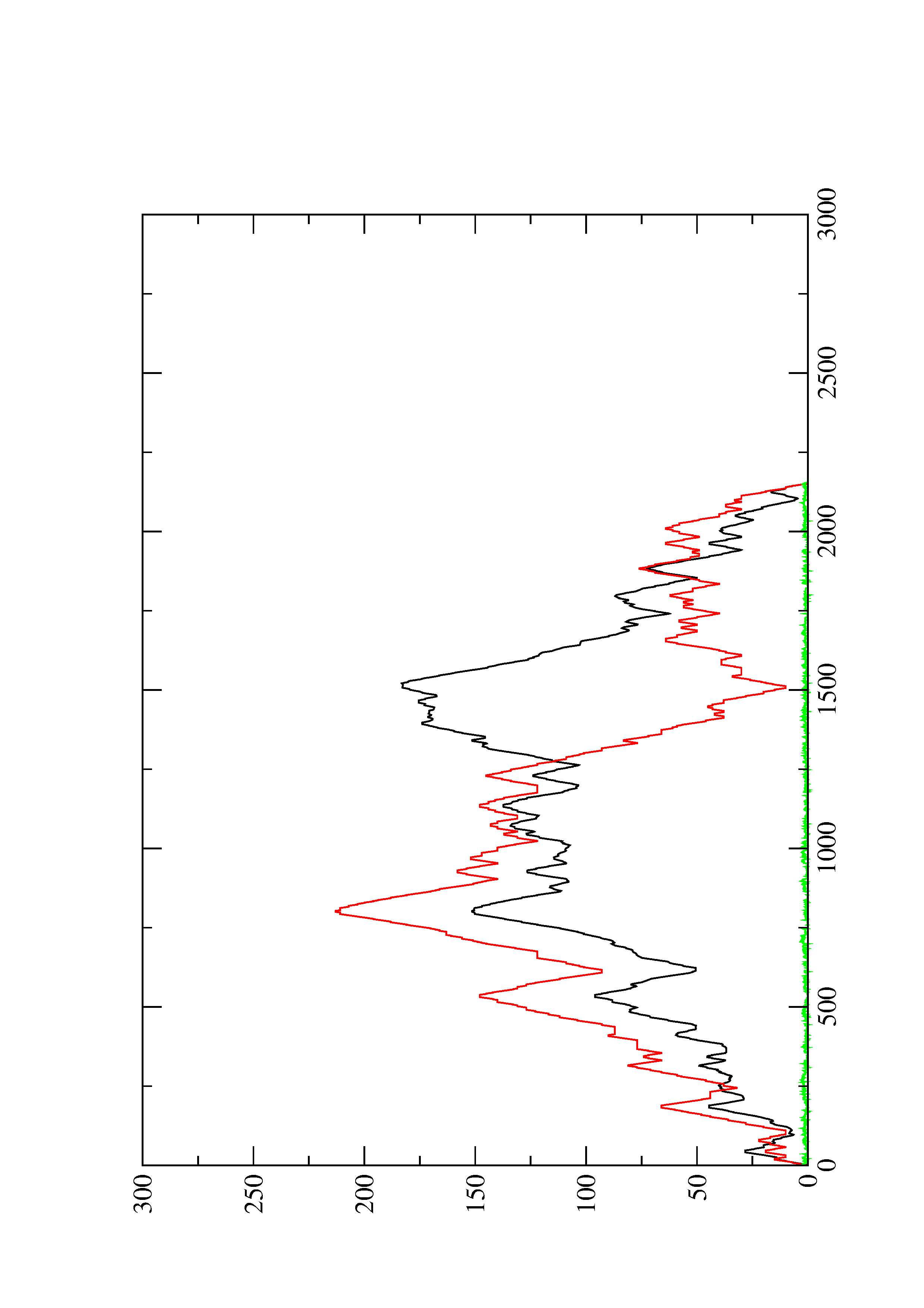
2. Here you find a mountain plot representation of the MFE structure, the thermodynamic ensemble of RNA structures, and the centroid structure. Additionally we present the positional entropy for each position. The resulting plot shows three curves, two mountain plots derived from the MFE structure (red) and the pairing probabilities (black) and a positional entropy curve (green). Well-defined regions are identified by low entropy.