lncRNA Structure Prediction
Species Name : Withania somnifera Sequence Id : GBHJ01007409.1
I. Result for minimum free energy prediction
The optimal secondary structure in dot-bracket notation with a minimum free energy of (-598.30) is given below.
1. Sequence-
CUCCAACCCAAGUCGUCCAACACCACCUCUCUCUCUAUCCCUUUUUCCUCUGCCUCAUUU
UUGAUUUCCACCACCCUGCCAUCGGCAAUGCCCUCACGGAAAGCCACAACCAUGAAAGUC
CCAAAUCCUCUCUAAUCUCUCUGACUCCUCUUUCUUCUCCUCUUUCUGUUGCAAUGACGA
AGACCCACAUAGUCGCGGGCGAAACUAGAUCCACAGAGGAGAUUAGGGAUUACUACAUAA
AAACCGGCUGCUGCAACUUUAUCUGCAAGCUUUCUAUAAGGAUGGAAAGUAGGUGAUAUC
AGUUUGGAGAAUUGCUAUUUUUUAUUAAUCAAGAAUUCAACAUCUUUUUGACAUUGGCUC
UUAAACCGUUUUUUUUGUUUGUUUUUUUCAUUUAUGUGUUUCAACAAUUUGUUAGUACAA
AAUUUUGUCUCUUUCUUUUUAGAUAGAUUGCAUAGUAGGAUACUGGUGUUUAGUGUUGCC
GAUUAAGCACUGGACACCUACAACAAUCCAGUUAAGCUCAAAUGAAAACUUUGGCAUGAA
UUCCACCUCUCAUUGGUAUAAUACUUGAUUUAAAAAGUGCUUCUUGUUGGCUUCUUUAGU
GUUGAACUUCUGCUUUACGCUACUAGAGAAUCUGAUAACUCUACUGAAAAAUGGGACAAA
UUUUAUAUAUUUCAGACAUUGAACUGCAGGAGACAAAAAUAAAUGGAGACGGUACCUUAA
UAAUUUUGGGGAGUUAAGAGUCUUUCCCUCAUUAUUUUCUAUGUCUAAGACCAAUGCACC
UCUUGAAAGAACAAAAGACUUGAAAGAUUUGACCCCUGAUAGUAAAUUUUUCUUGUGUAA
UGUCCUCAGUUCUCAUGUGCAGCAGUUUCAUUGCAUAUGUGAUAAAUUUGGUUCAAUUUG
UAGGAGGGAGAAAGGUAUGUAUUACAUCUUUUUUGUUGGUUUAUGUUAUUCUCUUAGUCC
UGCAUUGUCCAAUUAUUUCUAUUGAUGCUACUUGUCAGUAUGUAUUUUGAUAAGUUUCUU
AAAGUCUUCAAAAAUAUAGUCGUAUCCACAUCUUGGUCCUCCAAAAAUGCGCUACUUUUG
AAGUAUCUGACAAGCAUCCUAACAUUUUUGAAAAGUUAAGAGCAUGGCUAGCUAACAUAG
GGGACAGAGGUAUGGCAAACUCAUCCAAACAUUAGUAUUUUGUAUCAUAUACAAACUUUA
UGAUAUUGCUCAGGCUCUCGAGGCAUAUUAUCGGGGCUUGACUAGAAUCGUAGAAAGAAG
CUAGGGAGAUUGCAAUCUUGCAUCCAUGUGGUCAGAUAUUAAUUUCUCUAUGUUACUAUA
UUGGUGUGAUUUCAUUCGCAAAAAUGAGUUAUCAGAUAUUACUAAUGACUUUAACAAAUG
ACCGAACCUCUUGUUGUAUUAAAAUACAAUAUACGCCGUUAGAGACGACCUUAUAUGGAG
GCUGAAUUAAUUUUGAGAAAUCACCCAACUGAGUUAAAGAUCACAAAGUUAGAGCACUUA
AAUAAAUAAUGUUACAGGUUUAGACAACAUAGGUGAUUGUAUAGGAUUGACUUUUAUAAC
AUAUUUUUCAAUGUUGUGCAGGCUUCAUAAAUAGAUCAAUAUGAUACUGAUGUUGCAAAU
UUAUAUUUAAUAUGCCUACCCAUUUUCACUAACGAAUCUUAAUAUUGUAAUAACUUCUUA
UAUGUCAACUAUGUUAUAUGUUAUAAGUUUAUUAACUAGCUAUUACUUCUCAGCCUGAAU
AAUUCUGUUUGUUUCACUAGCAUAUCUUGCUCUAAUUAAUCCAUUGGGUCCCUGAACCUA
UUAGUAGGAUCAUUAGGUUGACAGUUGAGUAUGAAUAAGUACUAAUAACAUAUUACAUGA
UCUACCUGCCUGUCUUUGUGCUGAUGGAUUCUUAUGUGGCCUUGUAUACCUUCUAAGCAA
CAUUUAUCCUUGGGUUGAUUGAUAGUUGAGCAUGUCAUUUGUUUUCUUGCACCUUAAGUG
AUUUUCCCCUCGUAUUAUUAAAUCUGGCUACGUUCUUAUCAGAUGUAUUGUUGUCUAGUA
CUAUUUUGAUGUCUAAUGAGACAUUACAGCCAUAUGUGAUUAAAGAUCUAAUGGUUGAUG
UAUAAGGCUGAUUUUGUUUACUUAUGUAUUAAAUGAAGCCUAUUGUUGACUGACUAAGGC
AUAUGUGUUAAUCCCUGUUCCAUUCCAGACCCCUCUAACCUAUAAUAUGAUAAUUAUACA
UGAUUGAGUAUGACUGGUCCAUGCCCUUAUGUAUGUUGUCUUAUCAAUUAAACAAUCACG
AUCUCCCUUUAAUAUAGUUAAGCUUAUUCGUUGGUCCUACCCUCCAAAUGUAUGUUGUAU
UGGGCGAUUUGUAGACUAUGCAAACUUGUUUGAAAUUGGAAUAUUCUUAUUUGCUAAAUG
AUACCUGAGUUGACUAAAGAAAUGCUAAGUCCAACCUAACAAAGAAAUAAUAGAAUUAAC
UAUGGACUUGCCUAUCUUGUUAUUGCAAAUGGUCUUGCAUACAACUUUUUGUGCGUUCAU
CACUAGCUAUUGAAUGGAGUCUUUUAAAACUUGUCAACUUAUAUAUGUUCUAGGAUAGGA
UUUUAAAGUGAUAUGCAUGUUUCAAUGUGUAAACAUGAUGUUGAACUUUUCAAUAUGUUG
UAGUGUAAAUGUUUAAUGAACUGUUGCUUUUAUUUCAGUGAUUUCAUAAUGAAACCUGUG
GACGAACUUGCA
2. MFE structure-
........(((((((((((.((..........(((((...((((....(((..((((.((.(((((((((......((((...))))....((....))..((((....(((((.(((.......((.(((((((..((((((((((((((....(((....((((..((((...(((.............)))))))..
))))..))).....))))))).)))))))...............((((...((((........)))).(((((((((....))))))))).(((((((((.(((((((((.(((((((..........(((((((.........)))))))....((((((((((((((((((...((((((((((.((.((..((((.(
((((...((((...(((.(((((((.(((((....(((((((.((((..........((((...(((((((((((((........))))))))))))).......))))......((.((((.......)))))).........(((.......)))....(((((........)))))....(((((((.(((((((((
((((((.......)))).)).))))))))).))))))).)))))))))))..)))))))))))).)))....))))..))))).)))).)))).))))))))))))))))))).((((((.....))))))..)))))))))....((((....((((((((((((.((.(((.(((...((((((.....(((((.((.
.(((((((((.((........)))))))))))..))..............((.(((((((((........))))))))).)).....((((..(((..((((((((((((((.((((((.((((((.......)).)))).)))))).)))))))..)))))))))).))))...........(((((((((((((((..
......)))))))))..........((((((((...(((.(((((.......((((....))))..)))))))).)))))))).........)))))).......))))).....))))))))))))))))...))))))))))..)))).)))))))..)).)))))))........(((((((...))))))).((((
((((.(((.((((((((.(((.........))))))))))).))).))))))))....((((((((((((((..((((.((.........)).))))..))))))))))).)))(((.((((..(((((......)))))..))))))).....)))))))))............................((((((((.
...))))))))...))))))))))).)).))).))))).))))..........)).)))))))...)).))))....)))....)))).)))))((((.(((((((....((.((((..(((((((((((..((((((((...((((((((...((((((((........))))))))...((..(((...(((((((..
.)))).))))))..)).............((((((((.......(((......))).........(((((.......))))).(((((((((((.(((((((((.((((((((.....(((...(((..(((((((((((((......))))......((((.....))))(((((......))))).....(((.((((
....)))).)))))))))))).)))..)))...)))))..))).))))))))).))))).....................((((((((.....((((((((..........((((((...........))))))..((((((((((((((...((((((((.............))))))))......)))).)))))).
)))).)))))))))).))))))..((((((.....)))))).....((((((((....))))))))(((((((....((((...))))..))))))).......(((((...((((..(((....))).))))...)))))...))))))......))))))))..))))))))(((........)))............
....(((.((((((.((((((((....(((((.......)))))..)))))))))))))).))).......))))))))((.((((.((((((((((((((.((((......(((.........((((((.(((((.((((((((((((((((.....)))))).))))))))...)).)))))...)))))).......
..))))))))))))))((((...((.(((((((.............................)))))))))...))))......((...((((..((((((........))))))...))))...))))))))).))))))(((((((((((((((((........)))....)))))).))))))))...(((((((..
....))))))).(((..((((((((...))))))))..))).))))...)))))))....)))).)).))))))).)))).(((((...)))))..)))))))).)))))..
You can download the minimum free energy (MFE) structure in
II. Results for thermodynamic ensemble prediction
1. The free energy of the thermodynamic ensemble is [-651.05] kcal/mol.
2. The frequency of mfe structure in ensemble 6.77977e-38.
3. The ensemble diversity 801.38.
4. You may look at the dot plot containing the base pair probabilities [below]
III. Graphical output
1. A note on base-pairing probabilities:--
The structure below is colored by base-pairing probabilities. For unpaired regions the color denotes the probability of being unpaired. The reliability information of RNA secondary structure
plot in the form of color annotation, Positional entropy ranging from red (low entropy, well-defined) via green to blue and
violet (high entropy, ill-defined).
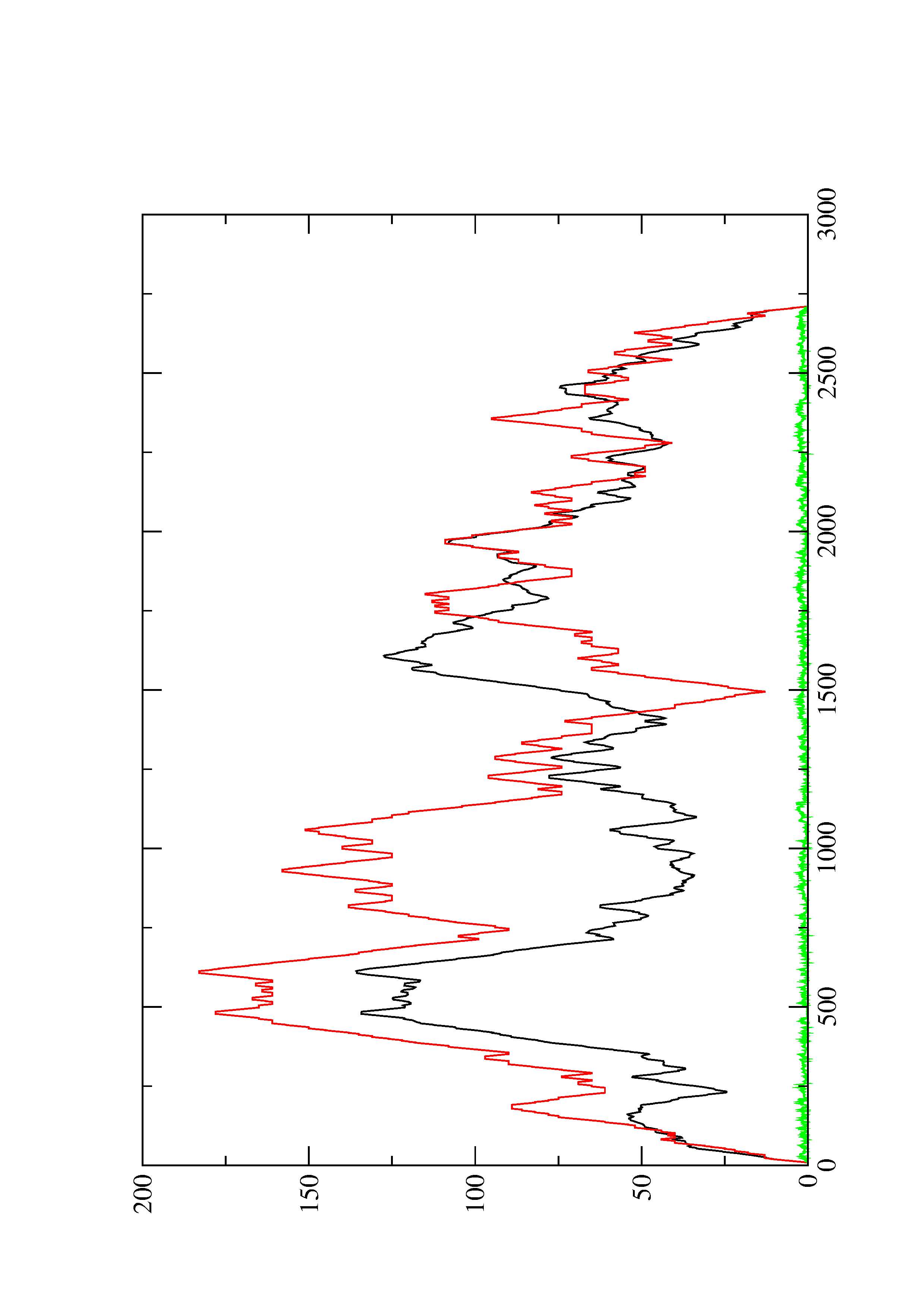
2. Here you find a mountain plot representation of the MFE structure, the thermodynamic ensemble of RNA structures, and the centroid structure. Additionally we present the positional entropy for each position. The resulting plot shows three curves, two mountain plots derived from the MFE structure (red) and the pairing probabilities (black) and a positional entropy curve (green). Well-defined regions are identified by low entropy.