lncRNA Structure Prediction
Species Name : Withania somnifera Sequence Id : GBHJ01008641.1
I. Result for minimum free energy prediction
The optimal secondary structure in dot-bracket notation with a minimum free energy of (-509.74) is given below.
1. Sequence-
CUCCAAUAGGAAUUCAUCUAAAUUAAAUAAAACAAGUUCUACACACUAAUAAUAUGAGAC
AUUAAAUAAAGACAAAGGAAGAAGUUGGGUUGAGACACUUAGGCCUUACUACUGACUACC
AAAUAUUUUUUUGGGUUAAGCCUGUUUAUUCUUUUGCUGUAUAGCUUCCUUUUCAGAGUU
UUUACGUAUGGCUCUUGGGCCAUUUCUUAUUUUCUUCGAACUACGGCUUGAAGAACUCGA
AGAAGUCUGGAUGCGAAGGAGGAUGAUGUUCAAAUUGAACUCGGAUGGAGGAGUACGGAA
AGAAAGGAGAGAAAAAUUAGUCAUCAAUUGACGUAAUAUGAAGACAUAAUCAAACUUGAU
GCAAAGCCAUUGUCAAUUAGAGCAUACAAAAGAAAAGCAAAAUAGCAAGAUCAUCACUCC
AGUUCAAACAAAUAUUUGAAUGUAAGACCAAAAGGUGAGAAAAAGAAUUUUAAAAGCAAG
UUAAAGCAAAGCCACAGUCCAUCGGCUUAUCAUUCCAGACUCUGUAAGAACAUUCUCACA
UAUAACUACCAAUACAUCUUCCAACUAUGAGCCUAUGCUGGUAUGAAAAUCUUUCUAGGG
CAUUUCAGAGAGAGACCUAGGCCUUUACUGUAGCUACGCCAGAAUUUGGACCUAAAAGGA
GUUAACCAUACUCCUAAGCAUAUCUUUGAUCACCUCAAAAUCAGUGGGAAAACAUAUGGU
AAUCUGGAUUAAAAAGCUUAUCAUUUGGCUAAGAAAAUCACAGACUUUCAAAUGGAUCGU
UAGCGACGACGACGACAAGCUAUGGUAAGAGCUUUCCCUCAUCCUUGGCCUUACGUCUUG
ACGUUCAUUAAAGGACAAUGACAAAUGCAUGGACAACUUAAGACUUAUACAGUAAAAACA
UUGUCGACUUUUAGCUCUUUAGGAAUGUAGACAUUAUUCCUAAAUGACCAUUAGAGAAAC
AGGUUAUCUACUACGUUUGACACAGGCAGAUACACCACAUAAAGAAGACAUGACUCGAAU
GAAGCAUCAGUUUUAGCAUUAGGUUCCACAUACUUGCACAGAACACAAAUGAAGAUCUGG
ACUAGAACAAUCAUAACCAAAAGGAAAUAGAGCUUUGAGACCCUUCACAUAGCACACAAG
GUUACAGCAAGUUUUACGACAACUUAGACCUGCUUUCCAAGGUCUUCAACUUUAGCAUAA
AAUAGUAGCUAGCUAAGAUGAUCUAGACUUACACAUAUUACUGCAGAUCUUGACCUUAAA
GGGUCAGUUGUAGCCUAAUUCAUAUGAUUGCAGAUAACUAAUCAACAAAAGACCUCAUAG
AAAGAAAACAAAGAGAAACAAAGUGUAAAGGAUGAGUCUUUCCUAUCAAAACAAGGUAAA
ACCGAGCAAAUGCUAAUAACAUAUCAACCCCCGACAAGAUUUAAAGAGCUGAAAUUCAUA
GAGUCUCAAACCAGCAGGUAUGAUACCAUUCAGACCAACACAAAGUUUCAGUUGUUAGAU
AUGAAUAAGAGGCUUAAGGCAGUUGAGGAUUAAACAAGACACUGGAACUCACAAGACCAU
GUUUUAACCAAGCCAGCCAAGAGAAACCACAAACAACCAAAACGUGCUUACUCAUUCCCU
UCAAAAUGACAAGGAUACGGCUAUUUUCAGAGCAAAAACAUAAGCAACUGAUUCCAUCUA
GUGAACAAAAGCUUCCUAUGCUAGGUUAAUAUACAUGUCUACACACGACAAGAUCCUUAG
UAUUACUGGGGCAACUUCAACAGAAAAGGAAAAAGAACCGAAAUUUCAUGCUUUCAACAA
CAAGGCACAAACAGAAACAAGAACAAGACCACUAACCAGCCUAUUUCGGUCGACUCCGUC
AUGUUUCAGACCCUUAAAAUAGUAAGACCACACUUUAAGUAAUACAUUUUAUACCAAAAC
AGCAAGAAAAGGUUGAGUUUUCUACCUUGUUGGGGGCAGGAAACUCGGACGACGACCAGA
GGCUUAUUCCACCACUUUGACUCCGACUUGACAGAAAAAAACUUUCAACUAGUGCCUCCG
AUUCCAAAAAAUUAGGAAUAUUUUACCUCAAAACAUCGGGGAUCCAGAAAGAUCCCCGUG
AAAUAAAACUUCGAACUCUAGCCAAGAAAUCCCAAACUUUUUGGGGCUCUCUGCCAAAAA
UUUACUAAAUGAAUAGUGAAGGUAUUUAUAGGAGACGAUUGGAGUGGAUUUAAGGAUUUU
GGGCCAUGUGGGAGUAGAGGUUUUUGGGAAUUUUUUAGGAAAUUCACUGAUGGAUUUCAA
AUUUUGCUUCCACAGCUGGAGCGUUCGUACUAAUUUGGAACGGAUUUUCGGAUUUUGUUG
AGAGAAAAUGCCACCUAGAUCGUCU
2. MFE structure-
.(((....)))....(((((..............(((((((..................................((..((.(((.((.(((((...))))).))..))).))..)).(((((......)))))........((((((.(((((.(((((..((((((...((((..(((((((((.((((((....)))
))).((((...((((((((........))))))))....))))((((((((......((((((((((((((.((((((((((........)))).........................((((.....))))....((((....))))((((....))))............)))))).)))).............((..
....))...)))))).)))).(((((((......))))))).....(((....)))...................((........)).(((((..........))))).....)))))))).)))))))))........................((((..(((((..((((...(((((((.(((...(((((((..((
....))..)))))))..((((((....(((..((...))))).....)).))))..((((((.......))))))...(((((.(((..((((..........))))..))).)))))....((((((((....((((........)))).....)))).)))).((((.(((((.(((((((((.(((.((((....((
((.((...(((......)))..)).))))....))))))).))))........(((((((.....(((.((..................)))))....)))))))..............((((((((.......)))))))))))))))))).))))....((((((((.....(((((...)))))))))).)))....
...........(((((..(((((...((((((((((((((..((((.......((((((...............(((((........((((((((.((....)).......(((.((.((....)).)).))).....(((((((((((((((.......)))))((((((........)))))).....((((((....
...........))))))..(((((....................))))).((((((....))))))))))))))))......)))))))))))))..................(((................))).......))))))((((........))))..........(((...))))))).))))))).....
.........................)))))))..))))).)))))))).)))))))(((.(((......))).)))........)))).)))))..)))).))))...))))))....))))).))))).))))))......)))))))....((((.((((.((((((.(((.((((..(((.((.(((..........
...))).)).)))....((((((...)))..)))....))))....((((.((..........))..)))).......(((.(((.......)))))).))).))))))...)))))))).....(((((((((((...((((((((((((...((..((((.....(((....((.(((((((................
..((((................................)))))))))))))...)))....))))..)).)))).......................))))))))..............((((.......(((((((((((((.((((....)))).))))))))).......))))...((((((.(((......((.(
((((((.(((...((((.....)))).........((((.(((((.........)))))....(((((.....((.((((((((......)))))))))).....................(((((((.........)))))))(((.....)))......((((((......)))))))))))......)))).)))))
))))).)).....)))...)))))).(((((((((((((.......(((((((.((((........)))).)))))))...)))))))))))))))))(((.((((((.(((...))).)))))).))))))))))))))...............))))).....
You can download the minimum free energy (MFE) structure in
II. Results for thermodynamic ensemble prediction
1. The free energy of the thermodynamic ensemble is [-548.39] kcal/mol.
2. The frequency of mfe structure in ensemble 5.7875e-28.
3. The ensemble diversity 667.33.
4. You may look at the dot plot containing the base pair probabilities [below]
III. Graphical output
1. A note on base-pairing probabilities:--
The structure below is colored by base-pairing probabilities. For unpaired regions the color denotes the probability of being unpaired. The reliability information of RNA secondary structure
plot in the form of color annotation, Positional entropy ranging from red (low entropy, well-defined) via green to blue and
violet (high entropy, ill-defined).
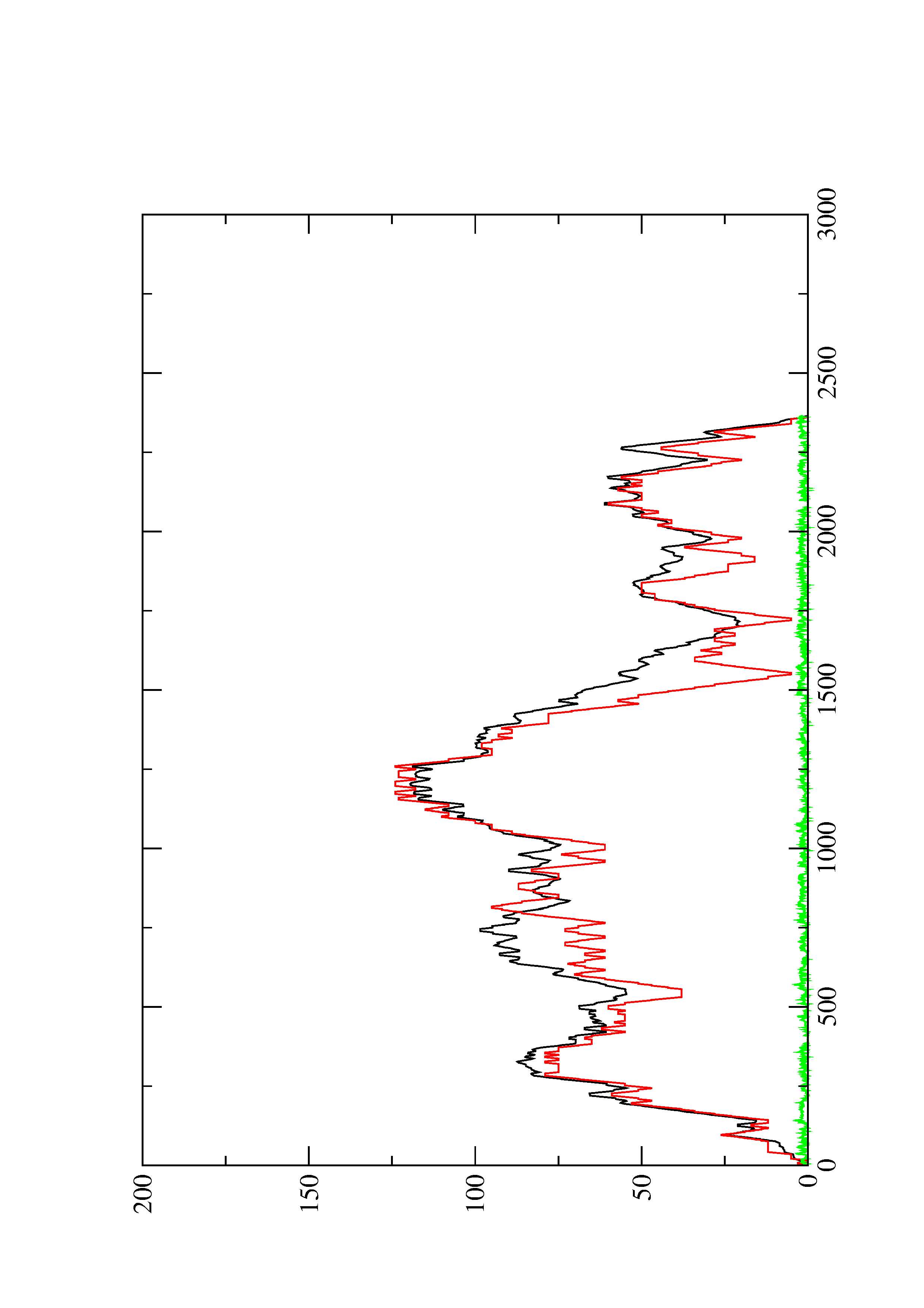
2. Here you find a mountain plot representation of the MFE structure, the thermodynamic ensemble of RNA structures, and the centroid structure. Additionally we present the positional entropy for each position. The resulting plot shows three curves, two mountain plots derived from the MFE structure (red) and the pairing probabilities (black) and a positional entropy curve (green). Well-defined regions are identified by low entropy.