lncRNA Structure Prediction
Species Name : Withania somnifera Sequence Id : GBHJ01011740.1
I. Result for minimum free energy prediction
The optimal secondary structure in dot-bracket notation with a minimum free energy of (-532.10) is given below.
1. Sequence-
CUUGUCUAUCUUUAAAUGAGAUACUAAAAAAAAUAAUUCAAAACCUCGUUCAACUAUGAA
CGUGGUAGGUAAUUAAAAUUUAAAAAAAAAUCAAAAACCCCGUUCAAGGAUGAAUAAGAU
UCUAAGACAAUCUCAUUCACCCAUGAACAAUAUACUCAUUAUGGGCUAACCCUAACCCUA
GUUGCCAUUUUGUUUUUUUUUUUUUUUUAAAAAAAAUGUUUUUGGGUCCUGGACUCAUAA
GUCUUUGAUGGUCUACUAUUCUAAUUUGAUCAUUAUGUUUACCAUCAAAAUUCUUCCUUU
AACAUUAAAUUGAUUGCUUCCAGCCAAAUUAAAAACUAUAGGAGUGAAAUAUUUGAAGUC
CCAUUUAGAAUUCAGUUCUUAAUACAUUAAUAUCUCCACAUUCCUAAACACCACUAAUUA
UCAUCCUUUUGAGGGUAAAUGUCAUAAAAGUCCUUUUUACAAAAAUCAUAAUGAUGGGAC
ACAUAAUUUUUCACUUCACAUUUUUAUUUAUUUUAUCAAAUCUAUAAUUCAAUACUGUAU
AUAUAAGACUCUUAUUAGCUUAAUAGGAAUAAUCUAGUUAAUAUCAACAUAAAUGGACGU
CCUUUCAUAUAUAGAGAGGAAGCAACAAUACAAUAUCAUUACAAGGAAGAAAAAAGAAAC
AUUGAAGUGAUACUACAUAUGUCAUUUCUGUGUUAAUACAUCACAUACUCUGCAACUUUA
ACUAAAAUGUUAGAGCAGUCAACACCAGGUCAAGAAAAUGAAAGGUACUGUUAAUAUUUU
CUUUUUCCAUUCUAAUAUAAGUGGACAAGAAGCCUACGUACUGAAUCCACUGGUUUUAAU
UUAAAUCCUACUUUUCCAUUAAGAAAUUCACUAAAAAUGUAUAUAAAUGCUUAUUUUAGA
AUUCAAUUAUUAAGACCUACAAUCAAUAUCUCAGAAUUUAAAUUCUGACAGCGCCUUCAA
UAAGAAAAUGCAUAAAGCACCUAACUAGCAAGAAAUAAUAGACAUGCCCCUAUUUGGUAU
GGGAAAGGUCAAAGUCCAACGAAAAAUUCAUCAAAAAGUUAAAAUUUUAUAAAAAUAAUA
AUUUUUAAUACAGUAAACAUCCAUUGAGUUAAAUCCCUUUAACAUAUAUAAGGAAAUAUA
AUAAUGCGUCUACAGAGAAAGUAGAGGGGAUUUUAACAUUGUUUUAGCCCAUAUAUUCAA
AUCUCACAAUAUUGUAGUAUUUUAGAAAAUUCUCUUUCUGCUAACCCUUCAUGCAAUUAU
AAACACCUCAUAAAUUAGAUGUGAAGAUAAAUGAGUGUAAAGUUAAUCACAGACAACCAU
UUUUUUUCCCUUUCCUAGUUGGACAUAAUAAUGGGUGCCAAUUAAACAGAAAAUAAAAAG
CAAAACCUUAUCUUCUCCUCUUGGUUGUCCCUUUUCAGUUCCAUCUCUUGUCAAUAAUUU
UUUUUUUUUUUUUCAACUUUGCCAAUUACAAACCACAUUUGCUUUAUUAAAAAAUAUGUC
ACAUUCAUUAUCACAUAAAUGGCCCUAUGUUUGGCUGGGUCAUGUAUUUGAUAUGACAUU
GUCCUAUAAUUAACUGUGUUUAUAAGAAAGUGCACAAAACAACUUUUUUGACACAAAGCC
CUUCCACUCCUGUCACUAUUAAUAGAUUAGAGCAAUUGUCCUUUUUAAUUAAUGAUACUA
GCUAGCUUCUAUAAAUUACAUCAAGCGAUUUACUGUUUUGUUGCAAAUUCACAUAUAAGC
UAGUACUAAUUAUUAAAAUCAAACUUGGUCCUUGUCUGUGUGUGACUAAUAAAUUCACUG
GAACUGACCCAAUCCUUUUACUUAACGGAUUUAGAUUAGAAUUACCUGUAGGCUUCUCAU
UCCAAUGAGAAAGUCAUAAAGAACUGAAACUGGAAGGAUAUAUAUUAUUUCUAAUGUUGG
CUCUACUAAUUUGAAUCUCAAGGUGUCCCCUAAGUCCCAAUCAAGUGCACCAAUUCUGUU
CAAUGGUUGUCAACCAACACAAGCUCUAGCAAUAGGAAGAUCGGAUUCAAGAUUCAAAGU
UUAUGAUAAUGAAUUCAUAGUCUACUUUGAGCUAUAUAGAUGAUGAGUAUGUAACAUAUU
UCUAUAGUUAUAUGAAUCCUGGUUAUAUUAAAGCGUAAGAGGUCAAAUUUGCCCCUGUAC
UAUGAGAACCCAUUUUAUCUUAUUCGAAAAAAGUUUAUCUCCCUCUUAAUUAAAUUCAAA
UAUAUUCUCCAUCCGGUAGGAUUGUCAAAGAUAAUAGACACUCAAUCCCACAUGGCCCCG
ACCAUUCGACAUACCAUGUCAGCGCUACAACCCAUUUUAGCCUACCCUUCCCUCCACUUG
UUCUUCCAUAUCUCUCUCGCUAGAUUUGUAUCUAGGUAUUUGUAACUCCUAGGUUAGAUU
GCUAACUUUGUGUGUUUGCUAAGGAUGACAAAUGGAGGAGUGUGAAGUGAUUGCAGGGCA
AACCUGCACAAUAUAAUGUUGUGAAGACUUCAACUUGCUCCACGCCCGCAUCCGGUUUCC
UUCCCUAGCUUUUUAUAGCAUGUAGGUAAAUGCAUGUCACAUAUAUCAUAUACGUAUCCA
AAACCAAAUUAAAGGGCUGCUAGCUACGACCACCUAUAUCUUCUGUAUAUAAAAGUAGAG
AGAAACGUUUAUCACGUGAAUCCACUACACCUAAUAGUGAUUCUGUUUGGAUUCUUCUCU
CUUUUUUUUUU
2. MFE structure-
...(((((((((.....))))))...................(((.((((((....)))))).))).((((((((..........................(((((.((.(((((.(((((.......))))).))))).)).)))))..............(((....)))......)))))))).....(((((((((
........))))))))).(((((((..((((((((....))))(((((((((..((....................))..))))))))).((((((((((((.(((..((((.((((((((...................(((((((((.(((....((((((((.(((((...))))).....................
........................(((....)))(((((................)))))............))))))))....))).)))))))))......((((..((((....))))..)))).......((((((((..((((((((((((((((.....(((....)))))))))))...........))).))
.)))..))))))))...))))))))))))..)))....))).)))))))))......(((((.(((((((((.......))))))))).)))))...............((((...((((((......)))))))))).......)))))))))))..((((.(((.((((......((((((..((((((........)
))))).)))))).....(((.(((.......(((((((((............((((......))))....((((((..((((....))))...)))))).........))))))))).....))).)))..((((((....)))))))))).))))))).........(((.....))).....((((((..........
.....((((..(((((((...(((..((.......))...............(((((((((........(((((((....)))))))...((((........)))).((((((....))))))......((((((((......(((((......(((.((((.((((((.((..(((...)))..)).))..........
.(((((((((((((((......((((((......))))))......((((((((..((((...(((.((........)).)))...)))).(((((....(((((..((((((((((((..(((...........(((((((.(((........))))))))))....(((((.((((((.((((..((((.........
.((((((........)))..)))..........)))))))).)))))).)))))...((((.......))))..(((.((((((((.........((((((.......(((((.(((.(((((((...........))))))).))).)))))))))))...............((((((...((((((((.........
.))))))))))))))..............((((........))))..)))))))).))).........((((((((....((((((((.(((.((((.((.((((((......)).)))).))..))))...))).))))))))....))))))))....)))..))))...))))))))..))))).....))))).((
(.......)))((((.((.....)).)))).(((.(((((((....((((((((((((((....))))))...(((........))).((((((.(((.(((((((.((((((.((((((..((......((((((.....(((((...((............)).)))))......))))))(((((...)))))....
.))..)))))).))))))(((((((((((((.((((((((((((((........))))))...)))))))))).((((((.((((.((.....)).))))))))))......))))))).))))...........(((((((.....................((((.....((((.((......)).)))).....)))
))))))))..........)))))))..))).)))))).((((((..................)))))).((((((...(((....))).....)))))).))).)))))....))))))).)))......((((((.(((((..(((............((((((....))))))......(((((....(((((((...
.))))))).....)).)))...))).))))).)))))).))))))))....((((((.....))))))...)))))))))))).))))))).)))).)))......)))))...))))))))...)))))))))...(((((((......)))))))...................)))...)))))))....)))))))
)))....)))....................(((((.((((((((..(((((..(((.(((((.((((.......)))))))))))).)))))..)))))))).)))))...
You can download the minimum free energy (MFE) structure in
II. Results for thermodynamic ensemble prediction
1. The free energy of the thermodynamic ensemble is [-584.76] kcal/mol.
2. The frequency of mfe structure in ensemble 7.84944e-38.
3. The ensemble diversity 647.27.
4. You may look at the dot plot containing the base pair probabilities [below]
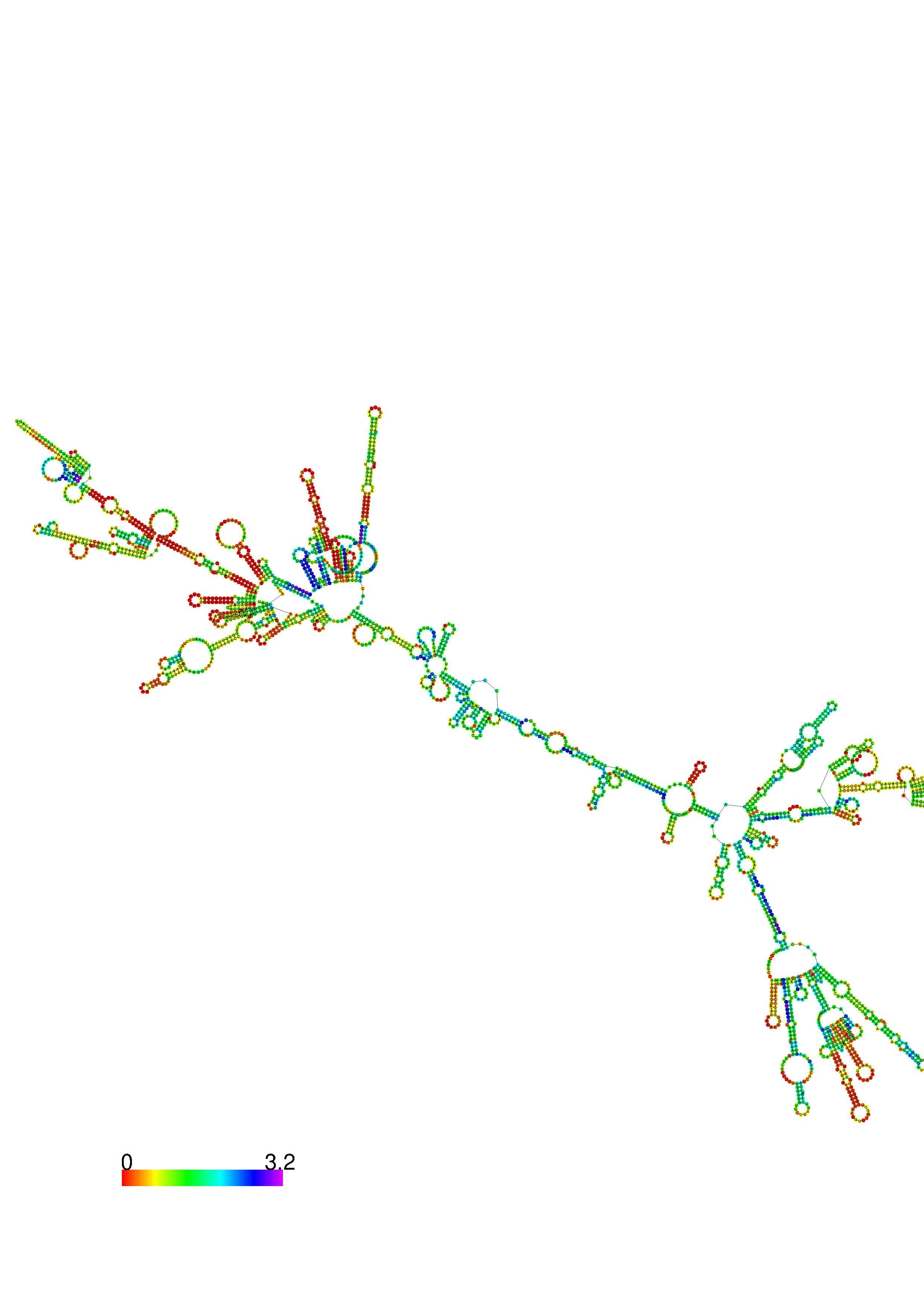
III. Graphical output
1. A note on base-pairing probabilities:--
The structure below is colored by base-pairing probabilities. For unpaired regions the color denotes the probability of being unpaired. The reliability information of RNA secondary structure
plot in the form of color annotation, Positional entropy ranging from red (low entropy, well-defined) via green to blue and
violet (high entropy, ill-defined).
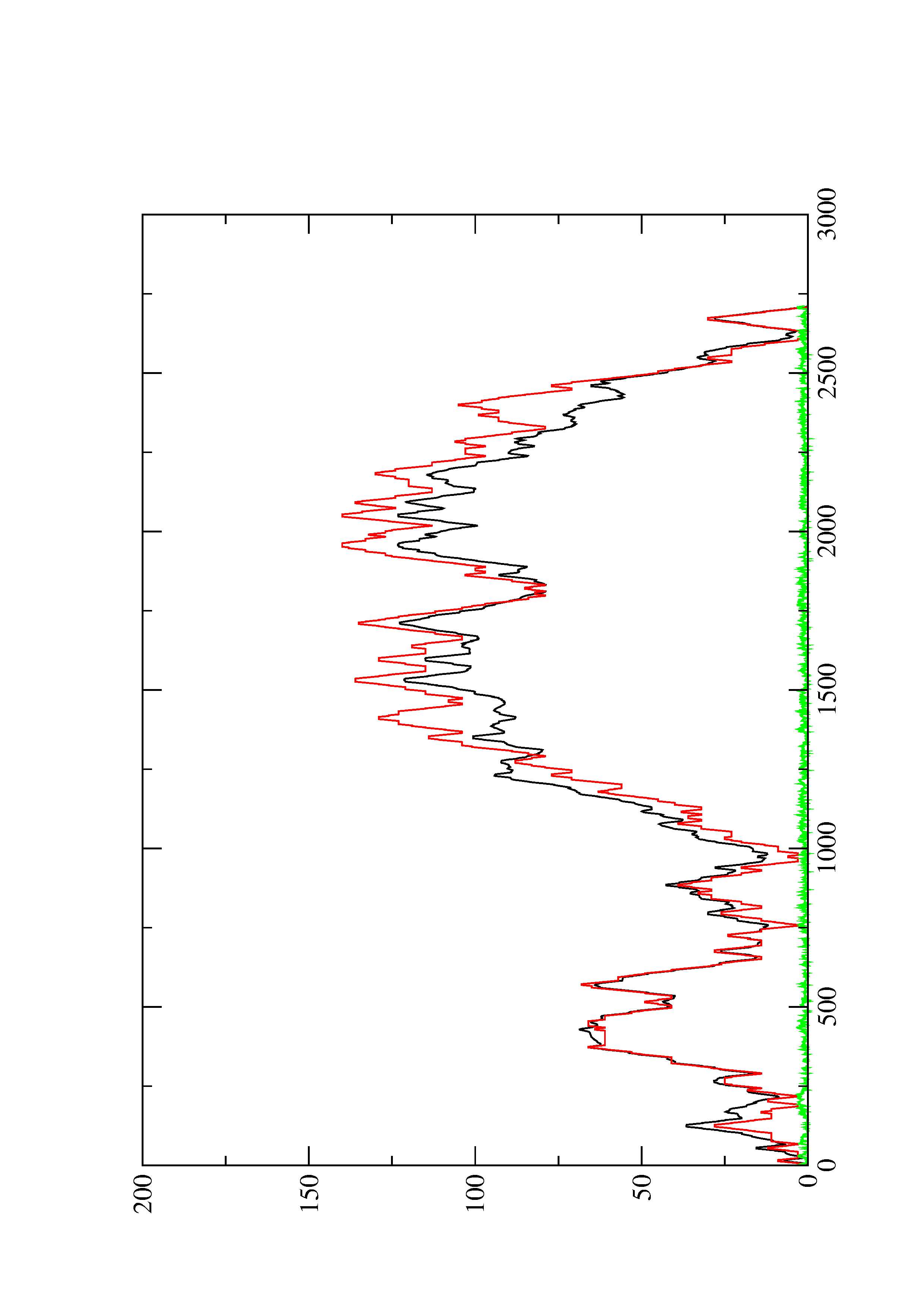
2. Here you find a mountain plot representation of the MFE structure, the thermodynamic ensemble of RNA structures, and the centroid structure. Additionally we present the positional entropy for each position. The resulting plot shows three curves, two mountain plots derived from the MFE structure (red) and the pairing probabilities (black) and a positional entropy curve (green). Well-defined regions are identified by low entropy.