lncRNA Structure Prediction
Species Name : Withania somnifera Sequence Id : GBHJ01012083.1
I. Result for minimum free energy prediction
The optimal secondary structure in dot-bracket notation with a minimum free energy of (-542.92) is given below.
1. Sequence-
UUUUUUUUUAAAAAGGUUAAAUUCUCAUUUUUAUUUAUUUAUUUCAUAGAACGCAGAGAU
UACAAUUCUUGGGUCUCACCAAAUUUCAUAUAUCGUAUAUCAAAUAGAUAUAAAGAAUUA
UAAAAAGCGAAAGGGAAAAAAAUUCCAAUUGAACCAAAUCCAGAACUCUGAUGGGCCAUA
GCCCGCAAGGCCAUGAAAAACGAAUACCAGGGCCCAAGUUACAUGUGCAAAAAGAGAAAA
GAAUUAGAAGAUGAUAUUAAUAAAUAAUACAAACAACUAUUAAGUCAAGCAUAAGUUGAA
ACUUAUAUUAGGCCAAGCCCAAGUGCAAAUAUACAUAUCAUUAAAGUAAAACACAAAGAG
GCCCAAUAAUAAAUUAUCAUCAAUUAAAGGGAAACAUGCUAAGAAAGGUUUCAUAUAGGU
CAAUUAGCUUAAUAAUGACAUGGACCUUCUGUUUUGAAAAUUUGGAAACUAAUCAACCUU
CAAAUAAUUCAACCUAUACAAACAAGUACUGUCCCAUUUAACCUUAAUAAAACAUGGCAA
ACCAAAUUUAAUUAUCUUAAUUAUACACAUAUUAUACAUAUAAGGUUAUUCAUGAACAUU
GGAAACAACACAAAGAUGAGAAAUUAUCUUAGUUCAAAGUGAGCAUGGAGAGUGUAGUCA
UAACAGAGUCAUCUUAGAGCAGACUUACAUAUUGCUGGUAAUGGAGAUAAACCAAUUAGG
UUUGCAUGCUAGCAAGUUAUGUGCUUGUGUGCUAUGUGUGACCGGUUGUACAAAAUAGGC
CUAUAACAAAUGUAUUGUCAUAUCAGGGCAGUCAUUGUAAGGUCACUAUGCAGCACCAAU
CUUACCAAAAAUUAUAUUUAUAAAAAACAAACUCAGACCUAGCAUGACUCAGGCAUCAAG
CCAUACACUACAAUAAACCGACAUAAGGAGGCAGCUAAACAAAGAACAAAGGAUCAGGUG
UUUGGGCAAUCAUACAAGUUGAGAUCAUAUAAAUGUAGAUCUAAUAUAGAAAACGGAAGG
GAGCAGGACGCAACAUAAAUUGUAUAAACCCAAGAUCAGAGCCAAAUUAAGAUGCAACAU
AAAUAUUUGUACUUUAAUAUGAGUAACGGAUAUGCAGGAAACUAUUGUAUCUAGUGAUAC
AUGGGCUAUGGAUUGAUGGGAGGAUGGGGCUGGCUCAGAUAAUGUCACAUAAGCUUAUCA
CAAAGAGGACACAAGCAACAUACUCAAGGGAACAUCAUGCAUUUAAUAUAAGAAAGAAAG
GAUGCAAAUAACCAGGUUGCAGAGACAACAUAAUAUCAGUAUAAAGUAAAAGGGAACAAA
GGACAUGUUUGUAAUGAAAUAACAUAAUCUUAGCUAUGAAAAAAGAAGAACAUGUGUUAG
UUUAGUUAACAACAAGCAGGGACUGAGGGGAAGGGAUCAUGCUUUAGGAAAAUAGUCACA
UAACACAUAAUUAGAAAUAUGUAAGUUAGCAAUCUUAGUUCCAGUUUAGGGUCUAGAUCA
UUAUCAAACAAGCAUUUAGAUACAAAUCCAAGUAUAUAAGCUAAGAAACAAAUCAAUAUU
GAUAGAACUACUAUUCAACAAAACAUACAAAAAUUAAGAACAUAGAAAAUAGAAUUAUAA
CUCAGCAUAGAGGGCCUAAUGUUUUGAUACAACUUAAACACAAUUAAUAGGACAUUUAGA
UGUAUUCUGUCCCUCUACCUUUAUAAAUUCAAACUUUUGGAUUCAUUGGAGCUAAAUUGA
ACCCCACACCAACUUAUGUUUCUCACCUUAGGUACUCUUUCAAGUCGUAAGACGCUGGGU
UGAUAGAACAAGCAAUUGAUUCCCAGGAUUAACCUCACAAGUCAACUCCCUUUUCUUGCA
CUUACUAAGGUCUUGUAUAGUAUAGGCAAAGGAGAAAAGAGGAGAACAAGGUGAAAACUU
UCACCUCUAAGUUAUUUUGAAACUGAAAUCAAGACUUUAGAAGGAUCACUUAGUCAUCAG
AGCGUACAUAACAGUUGAUUUGAAAAAAAAAUAGAGUGAGAGGUGGGUUGAGAGAUCUUU
GCUUUAUGACUUUGCGGUGUCAAAGUGACUAUAUGAAAGUCACUUUGAAAAUACAGGAUC
AAAUGGACCUAUGAUGGCCUCCCUUACCCUACUAAUCAUAUAGGCAACUUGAUUAUACCU
UUAGCACCCUUUUUGUUUUUAAAAAGGAAACUGGGGCAAAAAUUAUGAAUGAAAAGACUA
UUUUGACACAAAAAACACAGACAACUAAUAUUUAAAUCACAUAAUCCAUAUAAAGUCAAA
AUAGACACCCUGGCUUAAAUAACAUCAUAGACUACUGGGAAUAAUCCCAUUAGCUGGCAA
GAUAGGAACACCGAAUUUAAACUAAAAGCAUCCAUAUUGAGUAAGUAAAUUCAAAUGACC
AACAUCAACAAGUGUAUAAGAGACUCUUAUAGCAGACUUGCUCAUCACAGAACAUUUUAA
CCAAUAAAGACAUGAGAACUAGAAUUCAUAAUCAAAAAGUCAUCAAAUGAAACAAUAGCA
UGACUCAACUUGUUUCAAGCAUGUGCAAAAAUCAACACAUAACCAAAAUGAUCAGAGAUA
AGUUUAACCAGGAAGACCAGAGAACAUAACUAAGUUCACAGACCAUCCCCAGAAUCCCCC
AUAAACAGAGUAAGGGCUAAGCCAAGACUGACCAUGCUCACAACUAGGUCUAGUCUACCC
UGGUGAUAAGAGAAGGCAACAAACUACAUAGUCACUUUACUUUUAUCAAGUAAGGUGAAC
UAAAUGGUAAACAAACAACAAAUAAGAAA
2. MFE structure-
...........................(((((((((...............(((.((((((.(((...)))))))))....(((((..((((((............))))))..))))).......)))....((((.....))))...(((.(((..((((...((((.((.((((((((((((...((((.((.....
.......)).)))).....................................((((.((((((...............)))))))))).(((((..(((..(((((((((((((...))).((((((((((((.............................(((..((((............))))..)))......(((
..((..(((((.((((((((((.(((......))))))).....((((((((((((((.(((((((............)))))))..)))......(((......)))(((((((...((((((((........(((....)))....((((((...)))))).................))))))))((((((..((((
.((......((.(((((((....))))))).))))..))))..)))))).((((((.........(((((((.((((........(((((((((((((((.((....((((((.....)))))))))))))))))..)))))).(((.(((((.(((((((((((((..........))))....((((.((.((((((.
......))))))))))))..)))))).))).)))))))).......................................)))).))))))).(((.....))).)))))).................)).)))))(((((((..((........))...))))))).((((.......))))(((((.(((....))))))
)).......)))))))))))..((.....))........))))))))))).....))..)))....................))))))))))))))))))))))..))).)))))((....))..((((((....)))))))))))))))).)).)).))))..))))..))).))).....((((.......(((((((
(...(((...............))).((((.......(((((((...............))))))).((((.((((((((.............................((((((.......((((((((............((((........)))).........))))))))(((.(((.....))))))(((((((
((((..((((((((.......))))...............(((((((((........)))))..)))).((((.(((((((((((...(((((((((.............(((..(((.((((........)))).))))))................(((((((.....))).))))......................
...........(((((((((...((..(((((((((((.((((((((......))))).))).....)))).....((((.....)))).))))))).)))))).))))).....))))))))))))))))))))))))..))))....((((((((..(((((((.(((.......((((((((((....(((((...(
((((.((..(((....((((((..(((.....)))..........((((((((((((((.(((((((..........))))).)))))....))))))).)))).....((((((....))))))((((((...((((((........)))))).)))))).)))))))))..))))))))))))..........)))))
)))))......))).)))))))))))))))..)).)))))))))..(((.((((.....(((((((((((.......))))))))))).....)))).)))...((.((......))))))))))......(((......))))))))))).))))...((((((.((..((((((((.......(((..((((((....
.................((((((((((...............................................))))))))))....(((((.((((.........(((.(((.(((((....))))).))))))...((((.((....))..))))....(((((...((.....(((((((((.........(((..
....)))....((.(((((((...)))))))))..))))))))).....))...)))))............(((((........)))))........((((((...((......))...))))))..(((((((((...((((((..........)))).........))....))))))))))))))))))........
.((((........)))).........))))))..))).....))))))))...))))))))...(((((((((.............)))).))))).)))).))))))))....))))....((((..((((((((((((((.....)))))))))).))))..))))...........))))))))).
You can download the minimum free energy (MFE) structure in
II. Results for thermodynamic ensemble prediction
1. The free energy of the thermodynamic ensemble is [-600.04] kcal/mol.
2. The frequency of mfe structure in ensemble 5.66427e-41.
3. The ensemble diversity 669.13.
4. You may look at the dot plot containing the base pair probabilities [below]
III. Graphical output
1. A note on base-pairing probabilities:--
The structure below is colored by base-pairing probabilities. For unpaired regions the color denotes the probability of being unpaired. The reliability information of RNA secondary structure
plot in the form of color annotation, Positional entropy ranging from red (low entropy, well-defined) via green to blue and
violet (high entropy, ill-defined).
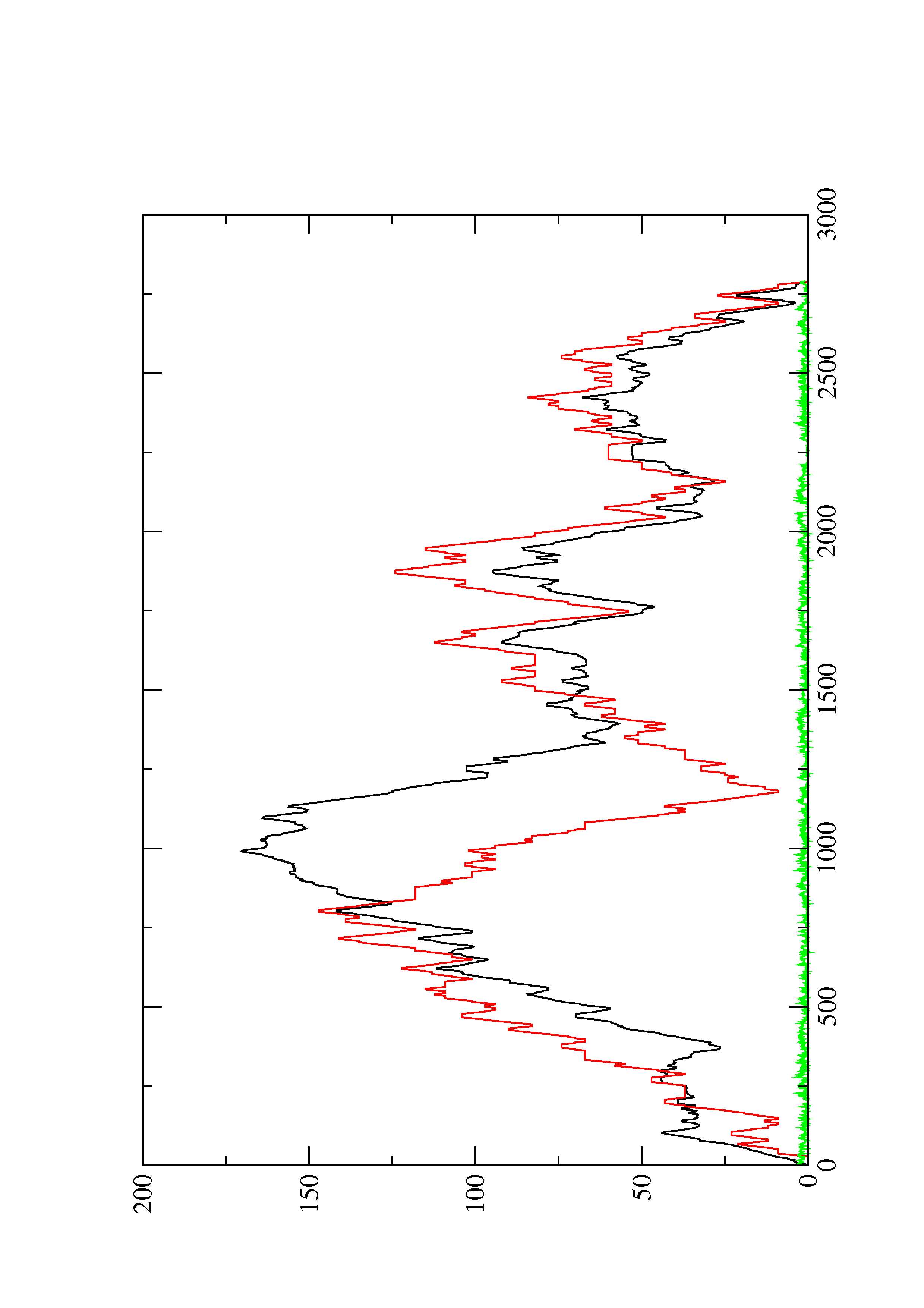
2. Here you find a mountain plot representation of the MFE structure, the thermodynamic ensemble of RNA structures, and the centroid structure. Additionally we present the positional entropy for each position. The resulting plot shows three curves, two mountain plots derived from the MFE structure (red) and the pairing probabilities (black) and a positional entropy curve (green). Well-defined regions are identified by low entropy.