lncRNA Structure Prediction
Species Name : Withania somnifera Sequence Id : GBHJ01012462.1
I. Result for minimum free energy prediction
The optimal secondary structure in dot-bracket notation with a minimum free energy of (-660.00) is given below.
1. Sequence-
AUCCUUUUCUAUCCGUUACUUCAUGAGAUGAACGAGUUUUCUACUGAAUUUCUCUCCAAC
GGUAAGGAUUGCAAUUUUCUCCUUGUUCCAGCUAUGAUUCACGUGUCUAUAGUACCAUUU
CGUACAACGUUGUUGUAAUUUGGUUGGUGGGUUGAAAUCCCUAAUUUUGAAUUUCAAAAU
CAGAAUCCUAGAAUUGAUUCUUGAUGGAUUGUACUAAGUUGCGUUUGAUGCUUGACGCAA
UCUUAUUUGAUAAGGUUAGAGUUGGUCUUCCCCAAAAUCUCCAUUGCCCCCAAACCCUAG
CUCUUAUCUCUAAAUAAUCAUAGUUAGAGAUAUAGGUUAAGGAGACAGAGCCGAGGGUCC
AUAUUAUUGAUUUACAAUAUUAGGUUGUUUUCCGGUUCUUAGAAGGAUUUUGGAGAGCUU
CAAGAUAAAUACUGGCUAUAUAACUUGGGGUUCAUAAUAGGAUUUAGUUUUUAGAAGGUU
UUUAAGAAGCUAGAUCUUGUUUUCAAGUCUAGAAUAUCAGUUUGUGCACAUAAGAGAAGA
AAAGAUCCUCAUAAACUUGUUUAAAAAUCUGAGUUCCUGCCUGGAUAAAGGCGAGAAACA
CUUAAAAUUUCAUGUUGUGAGUGCUGGUUUUUUUGCGGUUGGUAGCUUAGUUUGAAAUUU
GUGAACCAUUAAUGAAAAUAUCAAGUCUAAAGAGGAUGUUGUAGUGAUUGAAAGUAUGAU
CAUACAUGUUUUAAUAUGAGUAUGAAUCGUAUAUGAUUUGUGUGCCAUUAAAAUCUACCU
AGUUUUGCAUGAUUCUAAACGGCUAAGAUUGAAUAUCAAUUUAGUGUAGGUUUUAUUCUU
UUUUCAGUAUGAAUAGGGUGUAAAAGUUAGGAUCUUUGAAGGUAACUUGGUCAUUUGUGU
AUUGUUCACAGGUUACUUGAGGUUUUAGUAACAUGCUUAAGUUGUUUAAGUGGAGUUUGU
UGAUUUGUGUGCUGACUGUAUGUUUAAGUUAGUGAAAUUGAGUCAUGAUUUGUUGUUUUC
CUGUGUAAGCAAGGCUCAAAACUUGGAAGAUUUACUCUCUGAUGCCUUGAGUAAUCUAAA
AUAAGAACAGUAGGUUAGAGUCUAAGUCUCAGUAAUUUGCAAACAUGGUAUAGGUGAUUU
UGUUACCUAAGGUACUUUUAAGAUCCAAGUCUUGUCUAAUACGACAAAUUGGAUGUCCUA
UCUGUAGAGCAUGAUAUCUUAAUAAAACUAUUUUGGCUGUCUGAUAGCUUUAGAACAAAU
GCAUAUAGUUGGGGUUGUACACGUUAAGAAUUGUUAACUGUAGCUAGUAUAAAGAUACUG
UCAACAGUGGUUGCAUUACUUGAAAUAUGUCAGACCUGUGAAUUCCUCAUGCUGCUUGGU
CAUUGCUUGUUUAUUCAUAGGAAUCCGUUUACAUUUUUCUUAUGGGCAUGCUGAUCAAAU
UGUCCUUUAAUUUAAUAGAUGGGCAGUUGAAAUCAAGCUAUGAAACUUUUAUGGUUAAUA
GUGCCAUGAGAGAGCUGACAGUCCUGAUUUCUCACUUGGAAAGGAACCCACAUUUUUUUC
UUUCUCUGAAAUUCGACAUACUUAUCUCCUUCUAAACUUUCACUUGUUAAUCUGUGAUUA
CCCAAAUUAUAAGGUUAGACAUAGUCUUUGGUCUGUUGAGAGAAGGCAGAAGAAUUUAUA
UUGGGUCAUGCCUCAUUAUGUGAACUGUUGCAGGUUUGAAGUUAGUGAGGAUAACCUUCU
UGAUGGUUUACUUGUUUAAUGAUCAUGCUGCUUUAUUGUAUAUUUUCUAGUCAUCUUCAU
CUCUACCUAAACCUGGGCUGCUUGAAUACACAAUUAUAUUCGUUGAACUGGGUGGUGCUG
AAGUAAAAAAGAAUAAUAUAUAGUAGCUGAUCUAGGUGUAGUGAUGGUUUUGAACCAUUU
UUCUCAUGUUAAGGUUAGGUUUUUCUUGGAUUGUUACUUGCUAUAGUUGUUUGUAAAAGG
AGUAAAAAGUAACCUUCAAAUUGCAUCUGCUUAUUCUUGUUAAUGUACUCCCAUGCUAAA
UUUAUUUGUCAGGUAUAGUUGAGGUUGUUCUUUCGACUAUAGGGGUCGAUUUCAAUCAUU
AAAAUUGCUAAAAUAAUGAGAUAUUAGGUAGAACUGGAUUUGACUGCCUCGUUUUAGAUG
GGUCAAGUUCUAUCAUUUGAGUGUUUCUUCUUCAAACUGCCUGACUUUAUUAAUUUAUUC
UACAUUUUGCUAUUAUUUAACCGAGUGACGCAUGAAAUAUUAAUUCUUUUCUCUCUUUUU
GAAUGAACCACCUCUUCUGAGUCCCCUUGGACUUUCUUCUCGCAUUCUAUCGAGUCGAAC
CACUACGUCAAGCCUUUGAAGGAUGAAGACACAAGAAAUAAAGGAGAAACAGUGGGCUGA
AGCCCAUUAAUAGGAUAGCAGCAGUAACAAAACAAGAAAUGCGCCGAAGAGCUUUAGUGG
GGCAAGGCCCAUAAUCAUUUUGGUGAAGGGCUGACAGGCCCAUUUUAUCUAUGUUUAUUU
AUUUACUGGGUUUGUAAUAAGCCAUAUGUCCUAAAAAAAAUAUUACUUUCUUGCAUCAAU
UUUAUGGGCCACCUAGGCUCGACUUUUUUAGAAUUAUGAUAUGUGGAAGAGAAUGGGAG
2. MFE structure-
.((((.((((.((((.....((((((..((((.(((((((((.(((..((((((((.(((((((((((((((((((((..(((((((...((((((((......))).)))))((((.....((((((...))))))...))))(((.((((....)))))))..(((((...)))))(((((((((........)))))
.)))).........(((((((((((........))))))).))))...)))))))..)))))))((((((.(((((((((..............((.(((((..(((((((((.((....)).)))))))))..))))).))....(((((((((((.((.((.(((((.....))))).))))...)))).))))))).
....))))))))).))......)))).....((((((.(((((.(((((..((((((((((....((((..(((((((....((((...(((((.((((....)))))))))...((((((..((((.((((((((((...((.((((((.((.(((((((((((...(((((((((.(((..((.((((((.((...((
((((.(((.....))).)))))))).))))))))..))).)).))))))))))))).........................))))).))..)))))).))(((((((((((.(((((((.((((((.(((...((((((.......))))))..))).))))))..........(((.(((....(((((..((((((..
(((((((((......((((..((((.(((.((((((.((((.((((...)))).)))).)))))).)))(((((((.(((((((((..(((((((((((...((((..((((((((((.((((((((((....((((((.............(((((...(((((((..((((((((.........))))))))....))
))))).)))))..............))))))..))).))))))).))(((((((..(((((..((((...((..(((......)))))...)))))))))..)))))))..))))))))..))))...)))))))))))..)))))))........)).))))))).....(((((......)))))...(((((((...
((....))....))))))).......))))..))))..))))..)))))))))))...)))))..))).)))((((..(((..(((((...(((..(((((...((((((....))))))...)))))....)))...)))))....)))..))))..........)))))))..)))))))))))......((((....
))))..........)))))))))).))))))))))..((((((((((((......)))).))))))))....(((.(((......((((((((((.......))))))))))))))))..(((..((((((......(((((((((......)))))))))......)))))).)))...))))...)))))))))))..
..))))).....)))))...))))).))))).))))))...)))))))))..))))))))))))).))).)))).......((((((.((((((((((...........((((((.....(((((.((((....)))).))))).....)))))).....(((((.((((((.....(((((((((((......(((((.
...((((...((((((......((((((........)))))).....)))))))))).)))))......)))).))))))))))))))))))...(((.((.(((((((...)))))))...)))))(((..(((...((.(((((((..((((((((((((((.((.((((((((((((((..(((((...........
.(((((..((........))..)))))....(((((((((((.....((((((((...(((((((((.....((((((.....))))))...)))))....................((((((((.(((((((((.....(((((...((......))..)))))))))))))).....))))))))....))))..)))
)))))......))))))...............((((((....)))))).)))))...........)))))..))))))))..)))))).)).((((((((((((....))))))((..((((........))))..))........(((...(((....))).....)))...........))))))...(((((((...
.)))))))......))))))..))))))))..))))))).)))))....)))..)))))))))).))))))......((((((.((..((((.(((((((((...((..((........))..)).)))))))))....)))).....)))))))).............................(((((.....)))))
))))).))))...))))))....))))..)))).)))).
You can download the minimum free energy (MFE) structure in
II. Results for thermodynamic ensemble prediction
1. The free energy of the thermodynamic ensemble is [-712.08] kcal/mol.
2. The frequency of mfe structure in ensemble 1.99907e-37.
3. The ensemble diversity 742.98.
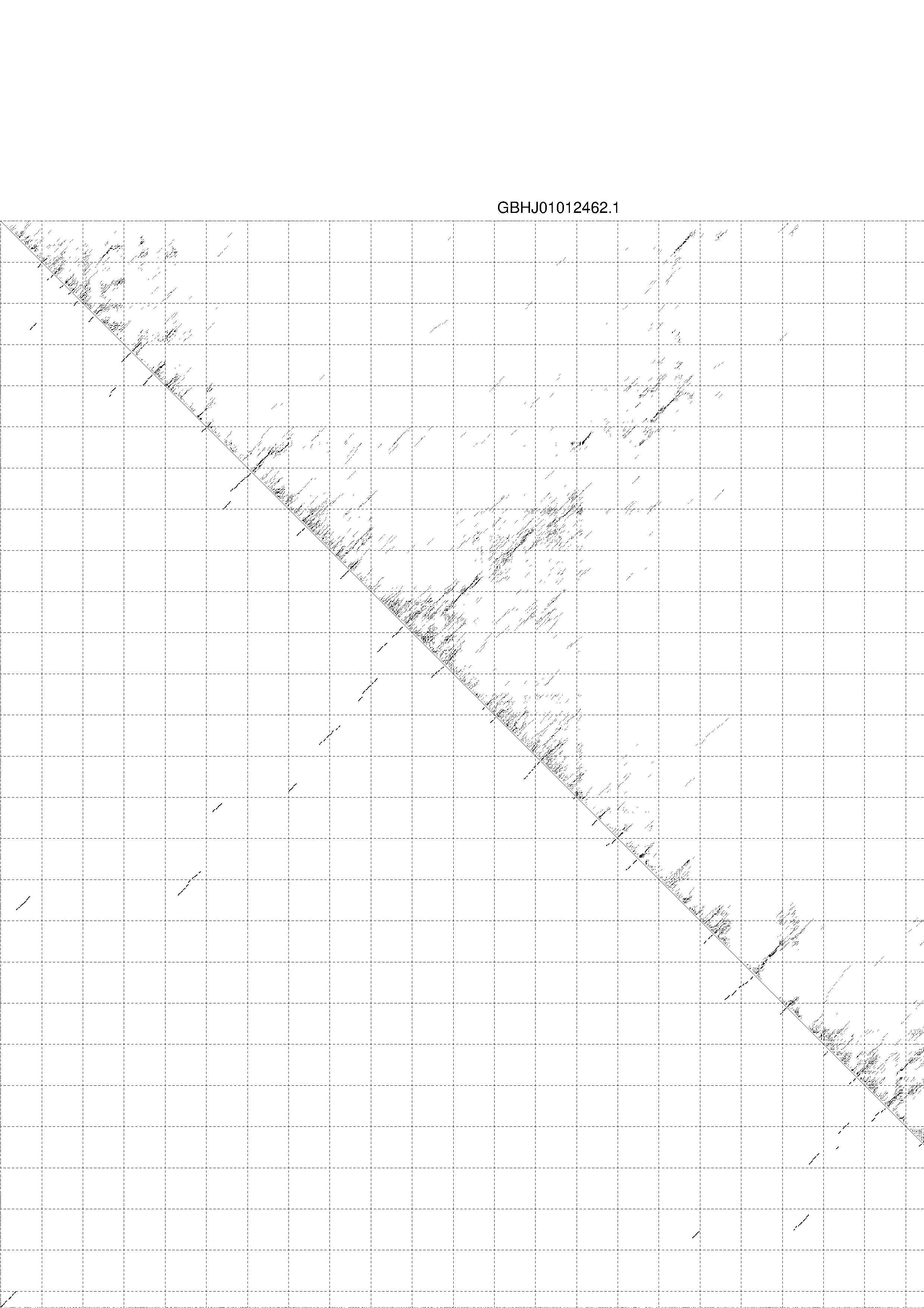
4. You may look at the dot plot containing the base pair probabilities [below]
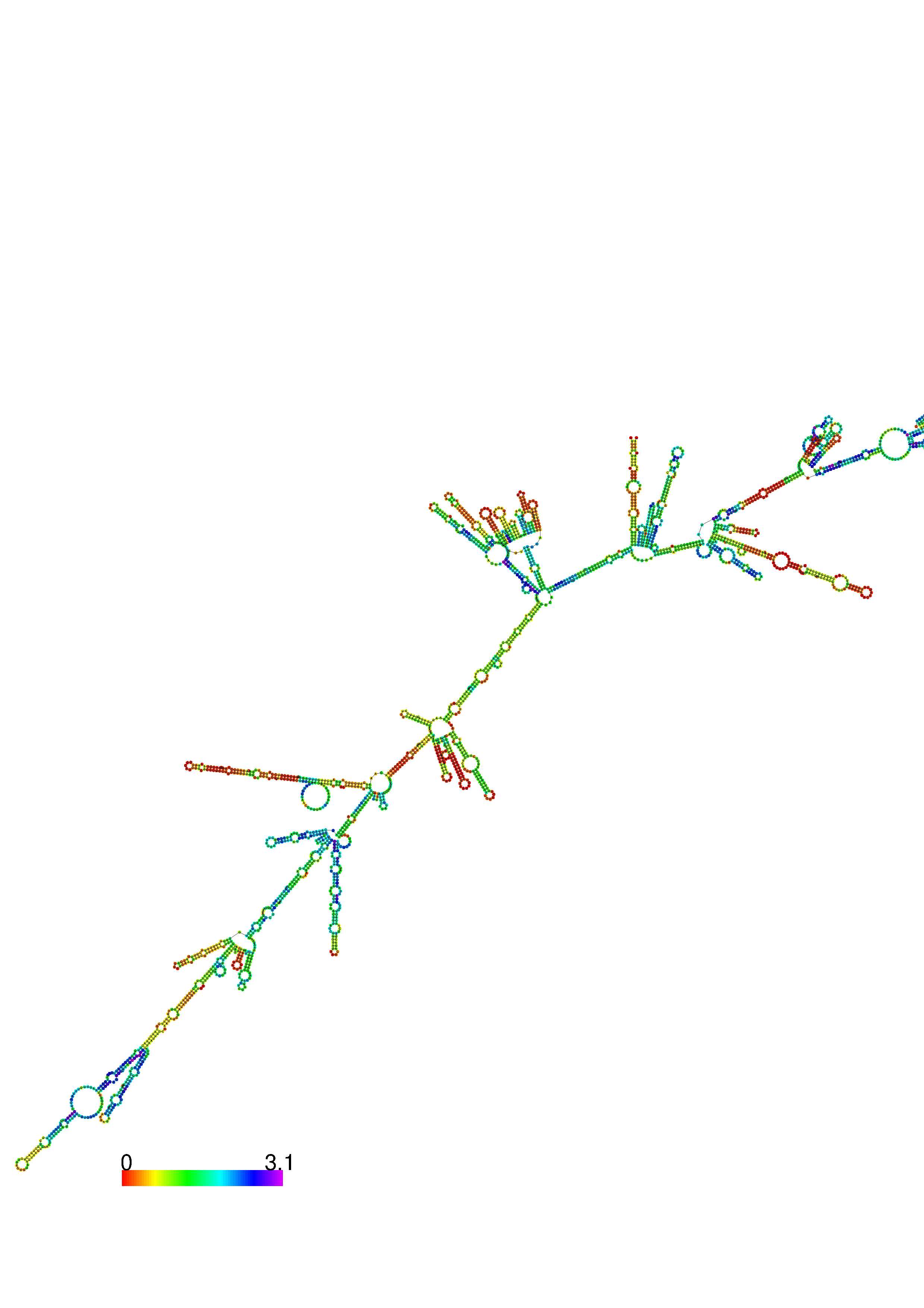
III. Graphical output
1. A note on base-pairing probabilities:--
The structure below is colored by base-pairing probabilities. For unpaired regions the color denotes the probability of being unpaired. The reliability information of RNA secondary structure
plot in the form of color annotation, Positional entropy ranging from red (low entropy, well-defined) via green to blue and
violet (high entropy, ill-defined).
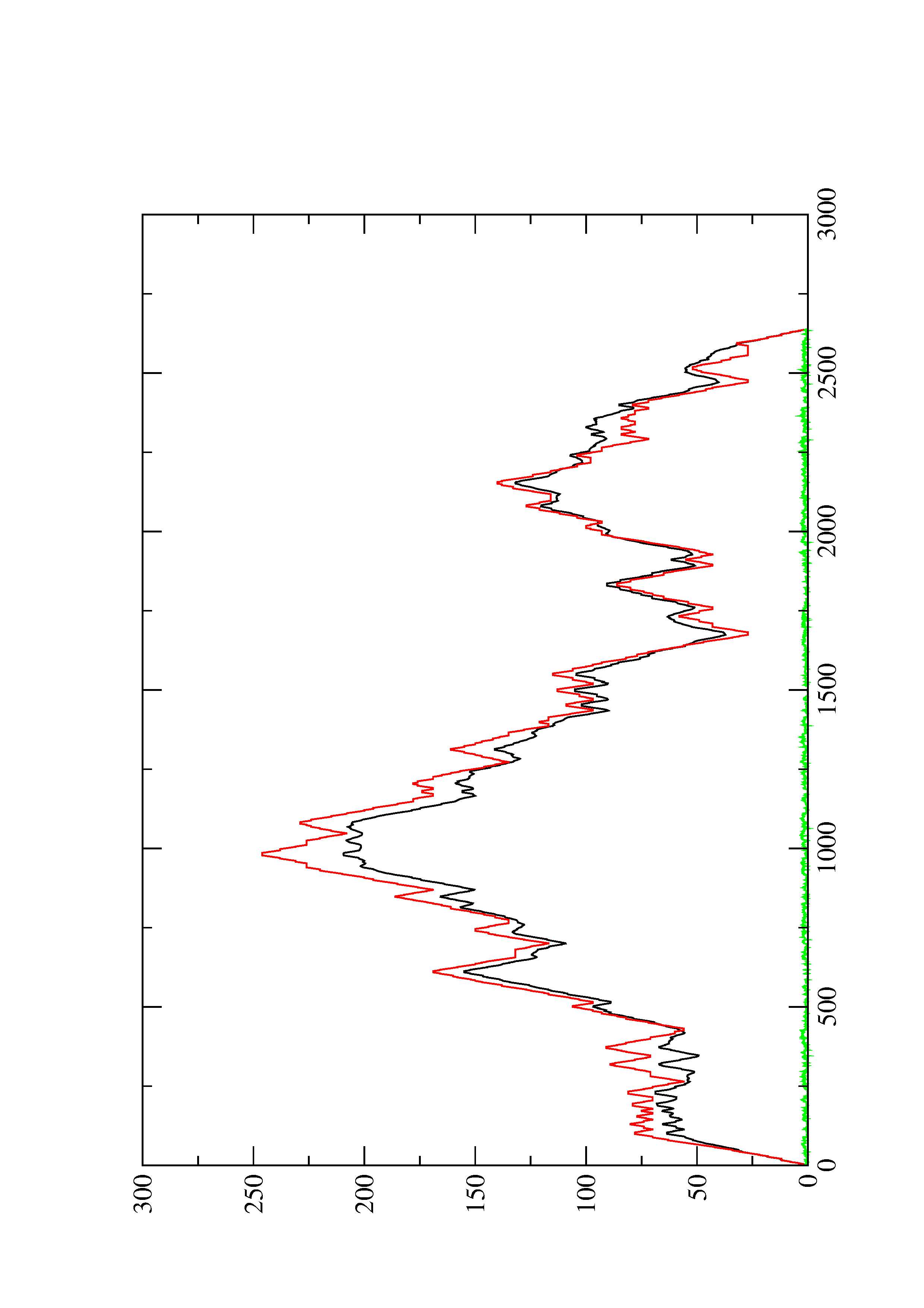
2. Here you find a mountain plot representation of the MFE structure, the thermodynamic ensemble of RNA structures, and the centroid structure. Additionally we present the positional entropy for each position. The resulting plot shows three curves, two mountain plots derived from the MFE structure (red) and the pairing probabilities (black) and a positional entropy curve (green). Well-defined regions are identified by low entropy.