lncRNA Structure Prediction
Species Name : Withania somnifera Sequence Id : GBHJ01012944.1
I. Result for minimum free energy prediction
The optimal secondary structure in dot-bracket notation with a minimum free energy of (-443.50) is given below.
1. Sequence-
AACAAGUUCAAUAAAUAAAUUCUAGUAACUCUACGAAGAUUAGUCUAAAUUUAAACAAAA
GAAUUUUACUCAAUAAUAUUACAAAGAAUUAAAUAAAGAGAAGGAAAGAAGUUGGGUUGA
GGCACUUGGGCCCAAACACCUACUAAUUUCCAACUUGUACUACCAAAUAUUGAUUGGGUU
AAGCCCAUUUUAUCUUCCGCUUCAUGGACUUUCCAGUAUAUUAGCUUCCCUUUCUGUAUC
UCACAUAUCGCUCUUAAGCCAUUUCUUAUCUUGCUAGCGGGGUUGACUCGAUAUAAUGCA
AAAGAGACAGACCAUUUGGACUCGGAUGGCAGGAGUUCAUGCAAAAGAAAAGGGGAAGGA
UUAGCACAGAUUAUUUAUUAAAGGUAAUUCCCAAAUGAAAAAUCAUUCUACGAAAGUGAU
GAACCAUUUAACUACGCAUGCUAUAGACAAUCAGUAGAUUUUAAAAGGUCAUAGGCUUUA
GCAAACACAAAAGCUAAAACGAUCAGGUUCUUAGCAAACUGCCGCAAAAAGAUUCAAUCU
CAAAUCUUAAUUUUUAUUUUUUUAUUUUAGAAGGCCUUUAUCUAUGUUCUUAAUCCACAA
ACCAUACAAAGUCUUUGCUUUUAAAAAAAAGAAUUCUGACUUACCGGGAAAACAAUGCAG
AAAAGGCAGACAAAUUCGAAGAAGAACCAUACACUAAUUGGCAUGAGGAUCAGACCUAAA
ACCUUCAUUGUAGCACACAUAGAAAAAGGUAAGGCAGUUGAAGUACAAAACAAACUAAAC
CAUACUGGAACAACAGUCAAAAACCAAACUUUUCAUUACAUAAAACAAACUCAACCAUCC
UAACAUCAUUUAAGGGGAGAAGGAAGCCGAACCUCUAAUCGACUAAUGACCAACAUUACA
CUUAAGACUUUCCAAAAGGCUAAUAGAAAGAGGAGCAGGUUUCAAAUUCAGACCUUACAA
CAGGGAUUAACAUAAGCAUCUUAAUUUUAGUCUUCCCUAGUCAUGCUACUAAGUGUUAGU
UUUAUACAAUCAACCUCUAGGCAUGUUCACUAUAUUCGGAUUGCCACAACAAACCUUUCC
CUACUCAGCGUGCAAGUAAUAGGACAUAGCACAACAAACAACAAAGUAGAACACAAUGGU
UUGAUUUCUAGUCUUGUUUUCAAGGCAAAACCAGCCAAAAGCAGCAAACAACAUGACCUU
CUGAGGCUACUUCAUUAGCAAAACUUUGAAAAUAAUGCAUAAACAAGAAAAUCCAGUUUU
AGCAGACUAUUGUGCAUAACGGGAUCAACAAUAUGUCUAUACUUUCACCAUUCUUAGAUG
CAAAGAUCUAAACCUCAAUACAUACUGAUCAGAAUAGUUAAUUCAAUACCUAAACAUACU
CCUGACCAUUAAAUGUAUGUAGACAACACAAAUUAUCAAAUUUCACUUAUAGAUUUGAAC
UUUAACAUUCAGAGAUCGAACCAAUUGACACCUUAACAUACUUUCAAGGCCUAAAUAGAU
GCAGACCCAUAUGACCAUCAUCACCUUAGAUACUUUAAAUUUAAACCUUUGACAUGAGGC
AAAGGUGAUAAGUUGGACUGAUUCGUUGAGGAAAGUUGAAGCUUUAACUGAACCUAAAGA
GCCAAGUAUACGGAUAACAAAUCACGAUCCAACAUACUCCUUGAUCCUCGUCCAGACAAC
UUGAAAAAAAGACGAGAAAAAAACAAAUACUUGCUUUCACAACAACAUGGCAUAAAUAGU
AAAAGUAAUUAACAUACUCUAAAUAGCCCUAUUCAGUCGAACAUACUCUGCCUAUUGCCU
UCAAAACCACAAACACCCAUUUAACAACCUAUUUUCUAUCCAAACAGUAAGAAAAUAAUG
GGAAUGAGGAAUAAUUCAAUCAAAGGAGCCAUGUUUACACUUUACAAUAGGAAACCACAC
GUUUAACAACCCAUUAUUUACCAACAAUGAAGAAAAAUAGAUGAGAGGAUGAGUAUUCUA
CCUGUUUUGGGGCAGGGAUCUGGGACGACGGCCUGAGUCUGAUUCCACCACUUCGCCUCU
GACUUGACUCGAAAUCAGAAUCAAGAAAACAAAAGUGCCUCCGGUUCCAAAAAAUAUAAA
UAUUAUACCCAAAGCGUCAAGAAUCCAGAAAAUUAUGGGAUUUUCUCCGUAAAAGAAAGU
UCUAGAGAAUAUUGAGAUAAUACCGUUGCUGUUGGUUCGUACAAUGAUGAUUCACCAGAA
AUAUCCUUUUGUUGAUUUUUUCCUUAACGAUUUGAGUCUGUAAGGGGAUGAGUACAGAAA
AGGAAUAUUGAGGCGGCUGCUUGUGAGAGUUUUAGGGUUCAUUUUG
2. MFE structure-
............((((.((((((((.((((((((((..((((((..((((((........)))))).))).))).......(((.............(((..(((((..(((.(((((..(((......)))..))).)).)))..)))))..))).............)))..(((((...))))).........((((
((((((((...))))..........((((.((((((((.((((....((.((((((.....((((((...((((...((((.....)))).......))))))))))(((((((..(((((((.........)))))))...........((((((((((((((((.............(((((((.(((.....((((.
.((((((.(((((..((((((((((.(((((((((..................))))...))))).)))....((.(((((...........((((((((......))).))))).(((((((.....((((((........)))))).........(((((......)))))((((((.(((((((.............
..........((((..((((((...........(((((......)))))............))))))))))...(((......)))(((.........))).((((((..............))))))........))))))).))))))..)))))))...............)))))))..((((......))))...
............)))))))..............((((((((..(((((((((((((((....((((((...(((((.......(((((.....)))))......((.((((....)))))).......)))))....))))))...)))...(((......)))...........((((((..((((..(((((....((
(.((((((((..(((((((((((((((.............(((((((..........((.(((......))).))...........)))))))..)).))))))).))))))............)))))....((((((((((((((((.((.((((((((((((........((........))...............
.....((((......))))....)))))))))))).))............((((.(((...(((........)))..))).))))........(((((((.............((((((......))))))......((((((..((.((((...(((...................))).)))).))....))))))))
)))))..........((((((((.......))))))))............)))))))))))).)))).........))))))...)))))..))))))))))........((((...))))..))))))))..............(((((((...(((((((((((((((.(((.((((((...(((((((..((((((.
...))))))...)))..(((((.((((((..((((...........))))................((((((..................))))))..........)))))))))))..)))).....(((.((..(((....((((.......)))))))..)))))....)))))).))).)).)).)))).))))))
)..............(((((.........((((((((............)))))))))))))..((((......)))).)))))))..((.(((.........)))...))((((.....)))).......((((..........))))..........))))..)))))).))..(((...(((((((((....(((.(
((((.....(((((...((((((((......((((..((......))..))))....))))).))).......)).)))))))).)))...................)))))))))...)))((((((.......(((((((((((.......))))))))))).......))).)))...((((.......))))))))
).))))))..))))...))).)).))))))))))))))))))))).......)).))))))))))))))))))))))))).))))...))))))))....)))).)))))))))))))).))))..
You can download the minimum free energy (MFE) structure in
II. Results for thermodynamic ensemble prediction
1. The free energy of the thermodynamic ensemble is [-488.74] kcal/mol.
2. The frequency of mfe structure in ensemble 1.31701e-32.
3. The ensemble diversity 480.44.
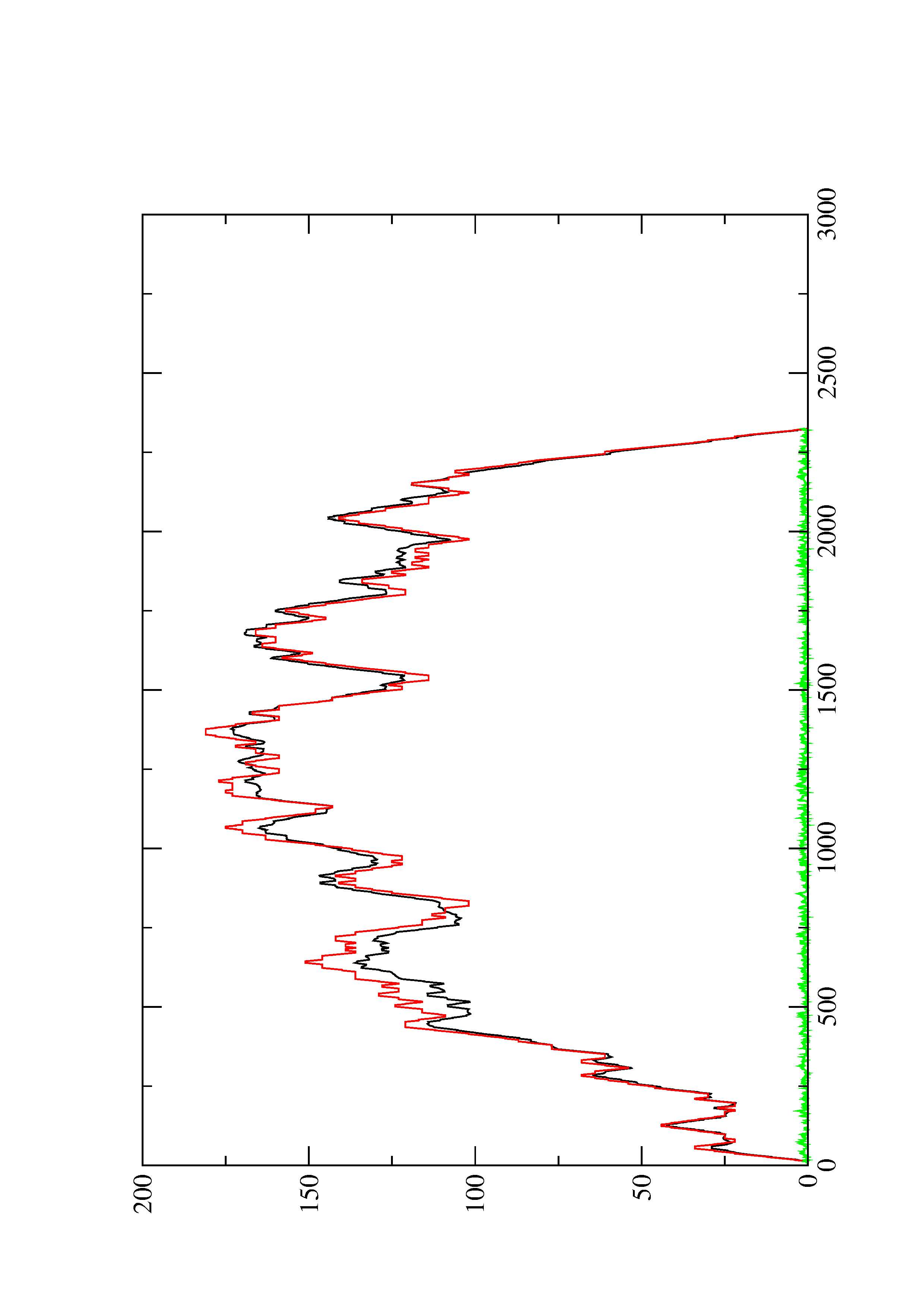
4. You may look at the dot plot containing the base pair probabilities [below]
III. Graphical output
1. A note on base-pairing probabilities:--
The structure below is colored by base-pairing probabilities. For unpaired regions the color denotes the probability of being unpaired. The reliability information of RNA secondary structure
plot in the form of color annotation, Positional entropy ranging from red (low entropy, well-defined) via green to blue and
violet (high entropy, ill-defined).
2. Here you find a mountain plot representation of the MFE structure, the thermodynamic ensemble of RNA structures, and the centroid structure. Additionally we present the positional entropy for each position. The resulting plot shows three curves, two mountain plots derived from the MFE structure (red) and the pairing probabilities (black) and a positional entropy curve (green). Well-defined regions are identified by low entropy.