lncRNA Structure Prediction
Species Name : Withania somnifera Sequence Id : GBHJ01013332.1
I. Result for minimum free energy prediction
The optimal secondary structure in dot-bracket notation with a minimum free energy of (-213.00) is given below.
1. Sequence-
CGGGGUCCGGGGAAGGGCCCGACCACAAGGGUCUAUUGUACGAACCUUACCUUGCAUUUC
UGCCAAAGGUUGUUUCCAAGGCUAGAAUGGGUGACCCACAUGGAAACAAAUUUACCACUU
UACAGUUGCCAAUAUGCAACAUAACUGAUUUUAAGUAAACCUGAUCUACUUAUGUAUGCA
AAUAAUAGAUGAUAGUAGAAAUUCCAAUCACAUAGUCAUAAUUAAGUAGUAACAUUCAUC
CCCUUACUACCCCUUUAAAGCAGUGAGGUGUGUUUAGUGUACUAAUCAAGAGAAGUGAGA
AAAAUAGUCCUACUUUUACUCAAGAGACUUGUCUUCUUAGGACAACAAAAAGAGGUCAGU
UUAGAAAAGACUAGAGGAGACUUGAAUAAAGAUUUGUAGAACAAUAAAGUGAAGGCAACC
AAAGUCAACCCCCAAGAAACUACUAGAAAGUAACUUGAUUUAAAGCAUAUGAGCCCAUUU
AACACUUUAAAACAUGAUACCCUACUUACUAUAACAAGAGGCCAUAUUCGUUAAGGCAAU
CUAAUGCUAGCCCUAUUCUACCUAACAUGCAUACUUUGCACAAAAACACACACGAACAAC
AACUAACCAUAAAUCUCUUAUCAAAAUCCCAACAAAACUAAGAUAAUACAGGGUGUAAUC
CCUUAAAAGACCUUAAGGUUUAACCUAGGUAGGUAGGAUGUAGCCAAUACUUAGACUAGU
AGCUAGACAGGUGGAACCGAACAAAUAACGACAUCUCUAAAAAACAAAAGGCUCAAGAUA
UAGCCUUCACAAUCCAUUAGAAAACUGAUAUUUCAACCAAGAUUGUUCUUGCAACAAGGG
UAAGUAACUAGAUAUACAAAUCAAGCUCUAGAAAAGGGUACUAUACACUUUUGUACAGAA
AAUACAUAACUUGAGUCAAGCAAGAUAACAAGCAGCAUCAAAAUCUGAUUUCUAUCGUGU
AGAUGUGCUUAGCAAACUCAAAUUUGAACAACCCAUUCAAAUUUAAAAGAAACAUAAACA
AAUAAAACUCUCCAUCAACAUUUUACUAAUUGAGAAAGUUGGGCACAAUCAGCUACACCC
GAACCAGUCCACACAAAUAAAGAAAAAGUUUCAAACAUAUACACAUACAAGUUUAAACCU
UGUCCAUUAAAGGGAAAAUAGU
2. MFE structure-
..((((((......))))))((((.....)))).(((((.........((((((((....)))..)))))((((((((.........((((...))))..))))))))................(((((......))))).....(((((..............(((((((((.(((.......))).)))..)))))).
.............))))).......(((((((...........)))))))((((((((.........((((((((..(((((........(((((((......(((((((..(((((.((..((((.((((((((...))))))........(((((..(((((......)))))...))))).......((((((((..
....(((((((..(((.......)))......((((..(((.......)))..)))).......((......))........)))))))............((.(((........)))))(((.((.....)).))).(((((.((((.((((((((((((((....((((.....))))....................
.................(((((...................)))))...((((......))))......((((..((((...))))))))))))))))).((((...((((......))))))))...(((((..................)))))...........((((((........))))))........(((((
....)))))........(((((....))))).....))))).))))..))))).)))))))).))))))))))))))).))))).))))))))))))..))))))))..((((.(((......)))..))))....((((.....)))).......((((((.((((((((((......(((((((((.......)))))
))))........................((((...................))))..)))))))))).....)))))).......................((((...))))..............(((((.......)))))...))))))))...)))))
You can download the minimum free energy (MFE) structure in
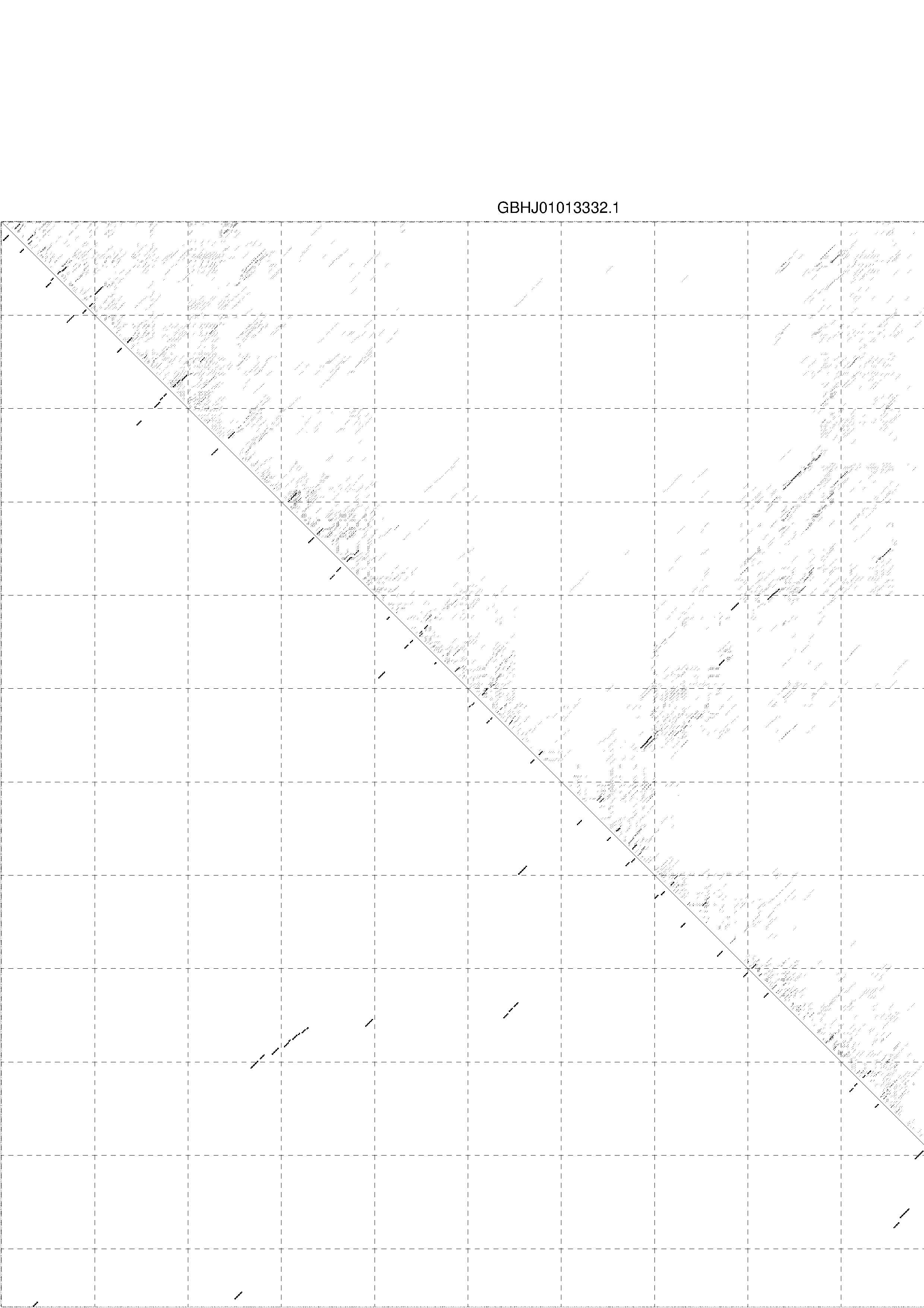
II. Results for thermodynamic ensemble prediction
1. The free energy of the thermodynamic ensemble is [-235.21] kcal/mol.
2. The frequency of mfe structure in ensemble 2.25147e-16.
3. The ensemble diversity 303.17.
4. You may look at the dot plot containing the base pair probabilities [below]
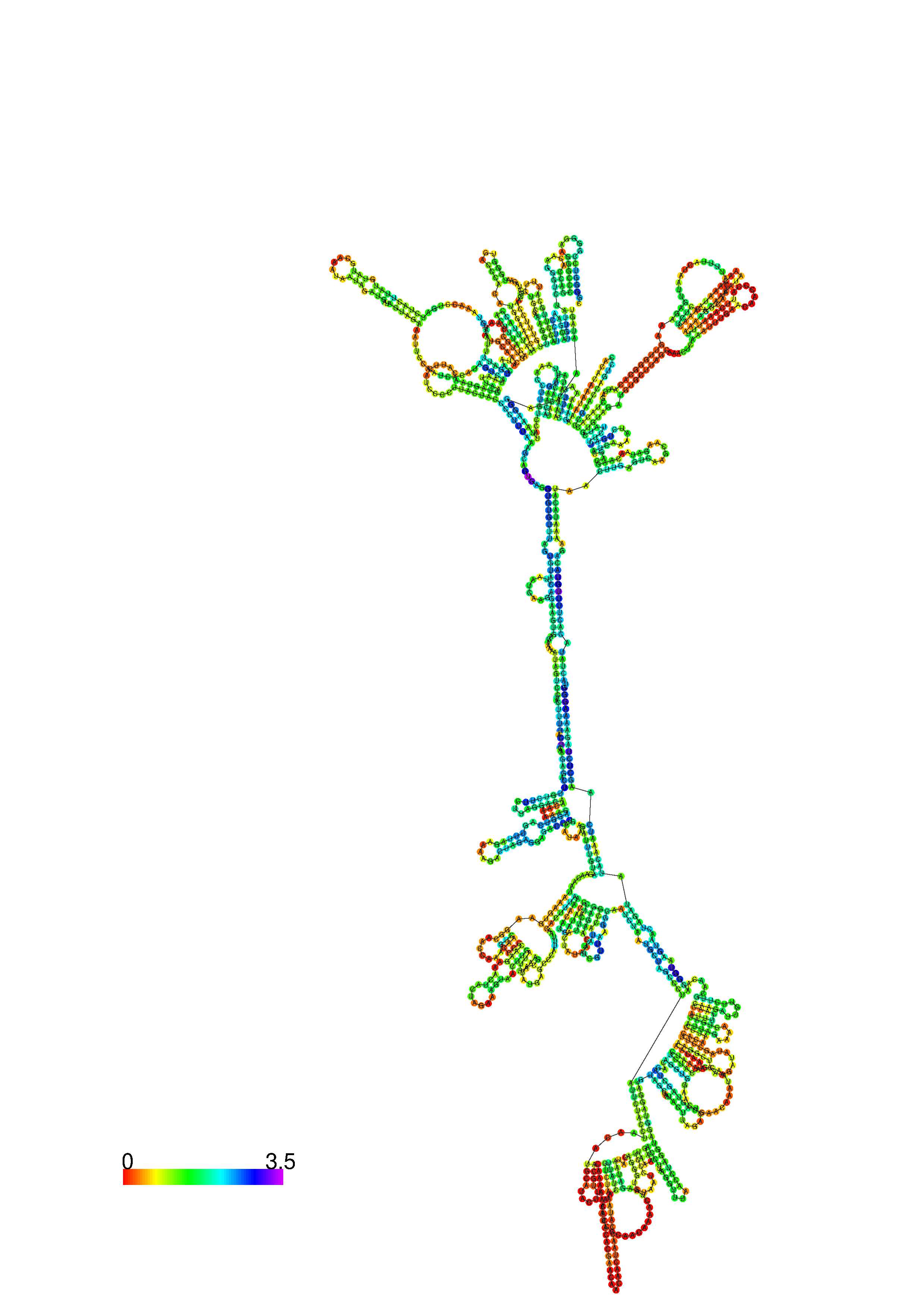
III. Graphical output
1. A note on base-pairing probabilities:--
The structure below is colored by base-pairing probabilities. For unpaired regions the color denotes the probability of being unpaired. The reliability information of RNA secondary structure
plot in the form of color annotation, Positional entropy ranging from red (low entropy, well-defined) via green to blue and
violet (high entropy, ill-defined).
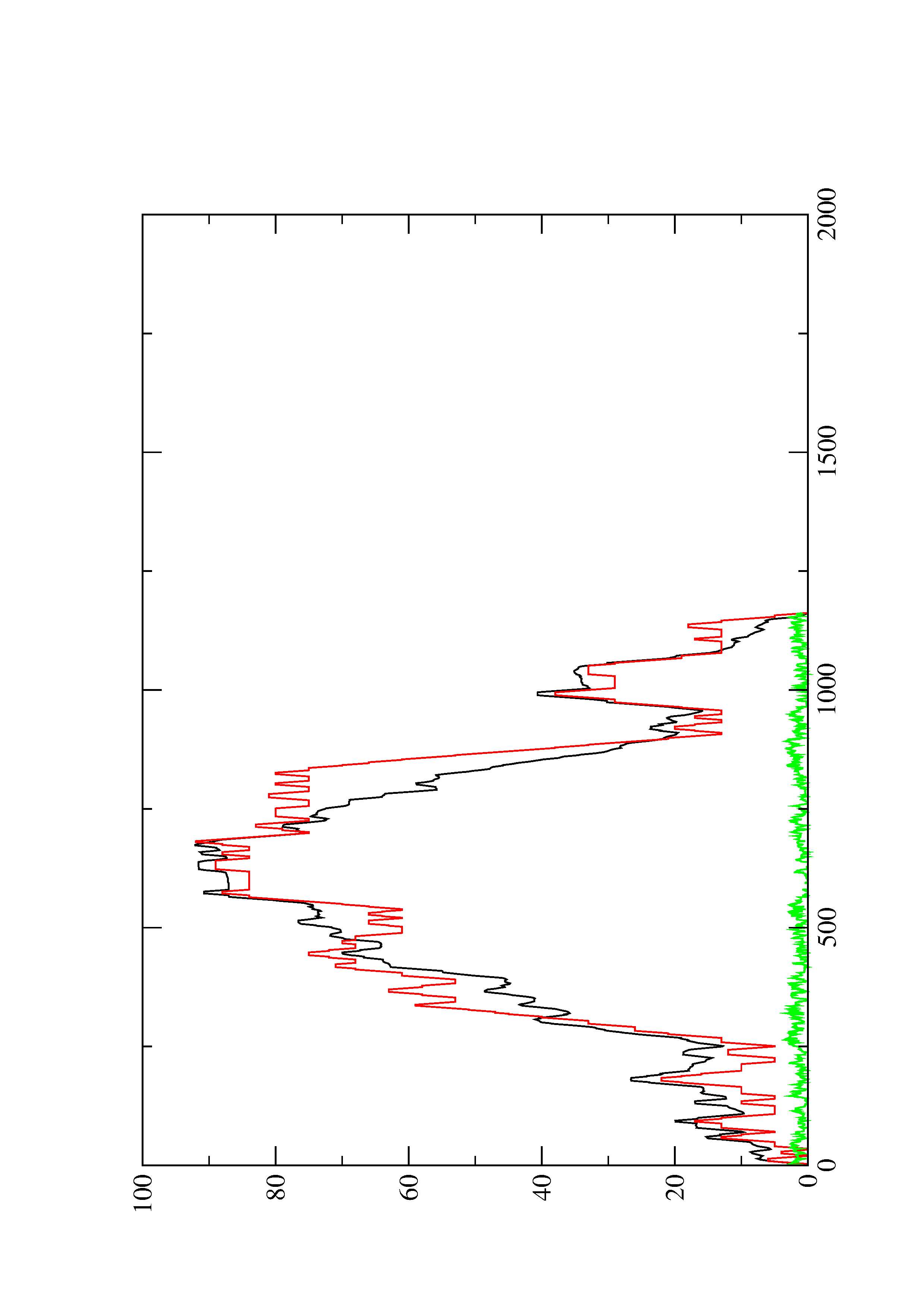
2. Here you find a mountain plot representation of the MFE structure, the thermodynamic ensemble of RNA structures, and the centroid structure. Additionally we present the positional entropy for each position. The resulting plot shows three curves, two mountain plots derived from the MFE structure (red) and the pairing probabilities (black) and a positional entropy curve (green). Well-defined regions are identified by low entropy.