lncRNA Structure Prediction
Species Name : Withania somnifera Sequence Id : GBHJ01013390.1
I. Result for minimum free energy prediction
The optimal secondary structure in dot-bracket notation with a minimum free energy of (-533.83) is given below.
1. Sequence-
AAAGGAAGGAAAAGUUUGGGCCUGAUGGCCUAUUACAAUCUCAGCACGAAAAUGAAAUAA
AAUGGGAUUUUGGAUUUCUUAAUGGGUUGCGGCCCAUUUGGAUUUUGCUAUUGCUGCGAU
GUAAAUGGUUUUCAACCCAUUUUUCUUUAGUCUUCAGCCUCCUAAGGAUUGACACGGGGU
CAAGACUCGACGUGGGCUUUGGGGACCAAUUGAAAAUGAUUCCAAUGGACUCAGGAUUGG
GGAGUUCAUGCAAAGAAAACGGAGAAGAAUAAAUAUCCAGCAAAUAAAUUUUUAAUUGAC
UUGCGACAUUCAAGGCUACUCAUUUUGAGAUAAUGUAGAAGAAUACAUGAAGUAUUUUCU
AUAUGCAAGUGAGCAAGUCCGAUUACGUAAAGACCCCAAGGAGAAUUAUUCAGGUCCCUU
GGACAUACUUCUCAGAAAGAUAAUCAUAAAGAAACCAAGCUUGAAUACUUCAACUCAUAC
UAAACAGAUCUAAACAUAAUGAUACCUUAAAAAUAAAUAAGCUUAAGAAAAAUUCUCGGG
GCAAUUCUGGUUAUUUCAAUCAAGUGAGCAUCAAACAGAAACAUUUCACAAGCAAACAAG
UCAGGCAAGAAGCAACAUGCUAGUUUCAACACCAAAAGGCAAAUUGGCCUCUAGUAAAUC
AUCCAAAGACUUCAAAUGACUUGCAUUUAAUCAUGUAGGCUUAAACAACUUUUCCUAAAG
AAUUCAACUUUCAACCUAGCUGCAAAUGAUUCAAGAGAGGCUAAGGUCAUGAUAGAUGAU
UCUCUGUUAAGAAAAGAUCUCAAUUUUAGCUAAUUCUCCAUUGUCAGACACUAAAGUGAU
CACAGUGGAAUACACAUAAAUGGAAUCAAUUCAGUCAAGAGGGCAAAAGGAAUAAUGGAC
CUUCUUGUUUCUCAUGUCCACCAAAACAUGUACAGCAUGAUUUUACUAUAAAAGUUGAGA
GAACACAAGGAGACCACUUCCUGCCAACUACUUUUCAAAAUUAACAUGCAUAUUCAUGGC
UUCAAAACAUGUAAGACUCUAACUAGCAUCAAGGCACUCCGCAAAAAAGAUGGAAUUUUG
GCAUGAAGACAGCAAGGAGUAGGAUCAGAUUUCAAACAAGCUACUUAAAUAUGGUUUUAA
CAUAUAAGACUCAUCUUAGAGACAUGAAAUAGCAGGUCUAAAAGAGAGCAAGUUACAAUA
CUAGACUAACAUCAAGGCAGAUUUUCUAACCAAAACCUAUUUACUAUCCUGAGCAAAUAA
GGCAUUCUAGGUCAAAACUCAUGGAUACCUAUUAAUAUAACUUAAAGAAGAGACAAGACA
CAGGAUUUCAUCAUACUUUCACAAAGACAAAUAAAUAAGGCUUUUGAAAAAAGUGACUGA
CGCCUAUGUUGAGCAAGAGUUUGCUUCCUUUCCCUACUGAAACUUUCCAAACACUCAAGU
UAUCCAUAUCUUAUUUUAACAUUCAUGCUGUGUUUCUUAGCGGAUUAGAAGCUAGGUGAU
UUCUAACAUAUUUAUCCUAAACAGGCAAUUAGAUCCUUGGACCUGCAUUAUUUAAUCAAU
GUGCACGCAGUAUGACGUUUACAGGAUUUUGCCAAAAGACUCCUACUUCAAACUUAAACG
GAGGUAUAGCAACAAAAAUUAGACACACACCUCUUUUCUCUUAUACUAGGUUAAAGACUU
GAGCCUUUCGACCAAAUCAAAGCAAAACUCUCCAUAACACUUCAGGACAGUAAAAGAACA
CCGGCUAAACAAAUAACAGACAAUCUCAAUCUUACACAAAAUAGAUUCAAAGUCCAUUAU
UAAACCAUCAAACACCCUAAAUCAGAACAGUCUACUUCUAUUUAACCCCCCUUAGCAAGC
UUUAGCCUUUAGGACUAGCCUAUGCAACUAUUUAUAAGCUAGAAAAAAAUAGAAAUAAAC
AAAGUCAUACCAGGUCACAAAAUUUAACAAGCUAAUCUUCAUCUUUUAACAUAUAUGAGU
UCAUUUAGUGGAUUAUUGAGCUAAACACCACACAAGCAUAGUAAUACACACACUGGAACA
GGGAAAUACGUGCACAAAAAGCCAGUUAUUUAACCAAAAUUUUGAGAAAAAUAAGAUGGA
GUAUAGUUUUUUUUUAUAGCUAUUUUAGGGCAGGAAGCUGGGAUGUUAACAAGUUGAAGU
GAUCUUCACUACCUUGAGCCUUGAUUUCAUAUCUUUUAAACUUCUACUAGGUUUAUAAAC
CAACAGAGAUAAUAAGGCUAAGAUCUGUGUUGGGCUCAGGUGAGAAGAGAUUCCAACAAC
GCCAUGGAUGCAACAACUACUAGAUGUUUGGAUCAUGCCCCACCAAGAUAGGGGUCAGAU
AACCCUAAACUCCUGAUUACUCAACUCAGACGGGAAUAAAAUUGACAAGAAAUCAUCAUU
CGCCUCUUUUAAGCCAUCGAUUAGGCUCACCCCCACAAUAUCCAACACCCCCAUUAGCAG
CAUAAAAGACUUGGGCUGAGGCAUUAGAAAAGAUAAGGAAAAGAGUUGAAAAACAUAAAA
ACUCCUUUGAAACAAUACAUGACCAACAACCAUCCAAACAAGCUCUUGCCACUACUCCCU
CAGUUAAAAUCGCUAAAAAAGGGUUGCUUGGGUUCUAUUAAAAAAUAGAGAAAUUAUUGG
CUAGUGCAGAGAGG
2. MFE structure-
.............((.((((((....))))))..))..((((.((((...(((((....(((((((.((((((...(((.(((((((....))))))).))).........((((..(((((...(((((((((((((((...((...((((((..(((((..............))))).)))))).)).)))))..))
))))))))((((((..((((((((((((((((.........))))))))........(((((((((....(((((.((((............((((((((((((..((((........((((...))))...((((((((((.(((.....)))))))))))))...)))).)))))).))))))........((((.((
(((((.((((((((...(((((................................))))))))))))).)))................(((..........)))..(((((............))))).....)))).)))).....)))).))))).......(((((................))))).......((((
(((.....((((.....((((((.............((((......)))))))))).............))))...)))))))((((((.(((((((((..............))))..(((.......))).((((((((.................)))).))))))))))))))).))))))))).((..(((((..
..)))))..)).....((((((((..(((((....))).))))))))))...........)))))))).))))))..((..((((...((((...(((..((((((((((((((........(((((((((...))))).))))...........))))))...)))))))).))).))))))))..))...........
....((((......))))(((((....(((((((((.((((.((........((....)).......)).)))).)))).)))))..)).)))...((((((..................))))))..(((((.......)))))....(((.((((..((((.((......)).))))..)))))))..(((......)
))......)))))..))))..........)))))))))))))..(((((((((.......(((.......)))....)))).)))))..................(((((((....((.....((((((.((((..(((((.((((..............)))))))))...))))(((((.(((((.((((((((....
)))))...((((......))))...((((((((.((.((((((((((........(((((((((..(((..(((.((((((.........)))))).)))..............(((((((..(((..((((((..((((((((((((............)))).....((((((((((((.((((.(((......))).
))))...............((((((.......................))))))....(((((....(((((((((.((((((((....((.....))........(((.(((...........))).)))...........)))).....(((((..(((......(((((..........)))))....)))..))))
)................(((((.((((........)))))))))...........))))))))))))).)))))(((((.((((........)))).))))).............)))))...))))))).))))).))).)))))).((((((..(((((((((.............((((.....(((..((((((.(
((............))).))))))..)))......)))).((.((((((..((.......))..)).))))..))...(((((....))))))))))))))..)))))).........)))..))))))).....)))..)))))))))...((((.(((..(((((..(((((.((((...(((.....)))...((((
(((.....)))))))....((((((.((((..((....))..)))).))))))))))))))).))))).))).))))......)))))))........))).))))))))))...........))).))))).)))))............(((((...............))))).............)))))).....)
)..))))))).((((........))))............(((((((((((...(((((.........((.((.((((((((...(((.........((..(((((((.....................................................))))))).)).)))...)))))))).)).)))))))....
))).)).))))))((((((....))))))....))))).....)))).))))..
You can download the minimum free energy (MFE) structure in
II. Results for thermodynamic ensemble prediction
1. The free energy of the thermodynamic ensemble is [-584.31] kcal/mol.
2. The frequency of mfe structure in ensemble 2.70597e-36.
3. The ensemble diversity 753.17.
4. You may look at the dot plot containing the base pair probabilities [below]
III. Graphical output
1. A note on base-pairing probabilities:--
The structure below is colored by base-pairing probabilities. For unpaired regions the color denotes the probability of being unpaired. The reliability information of RNA secondary structure
plot in the form of color annotation, Positional entropy ranging from red (low entropy, well-defined) via green to blue and
violet (high entropy, ill-defined).
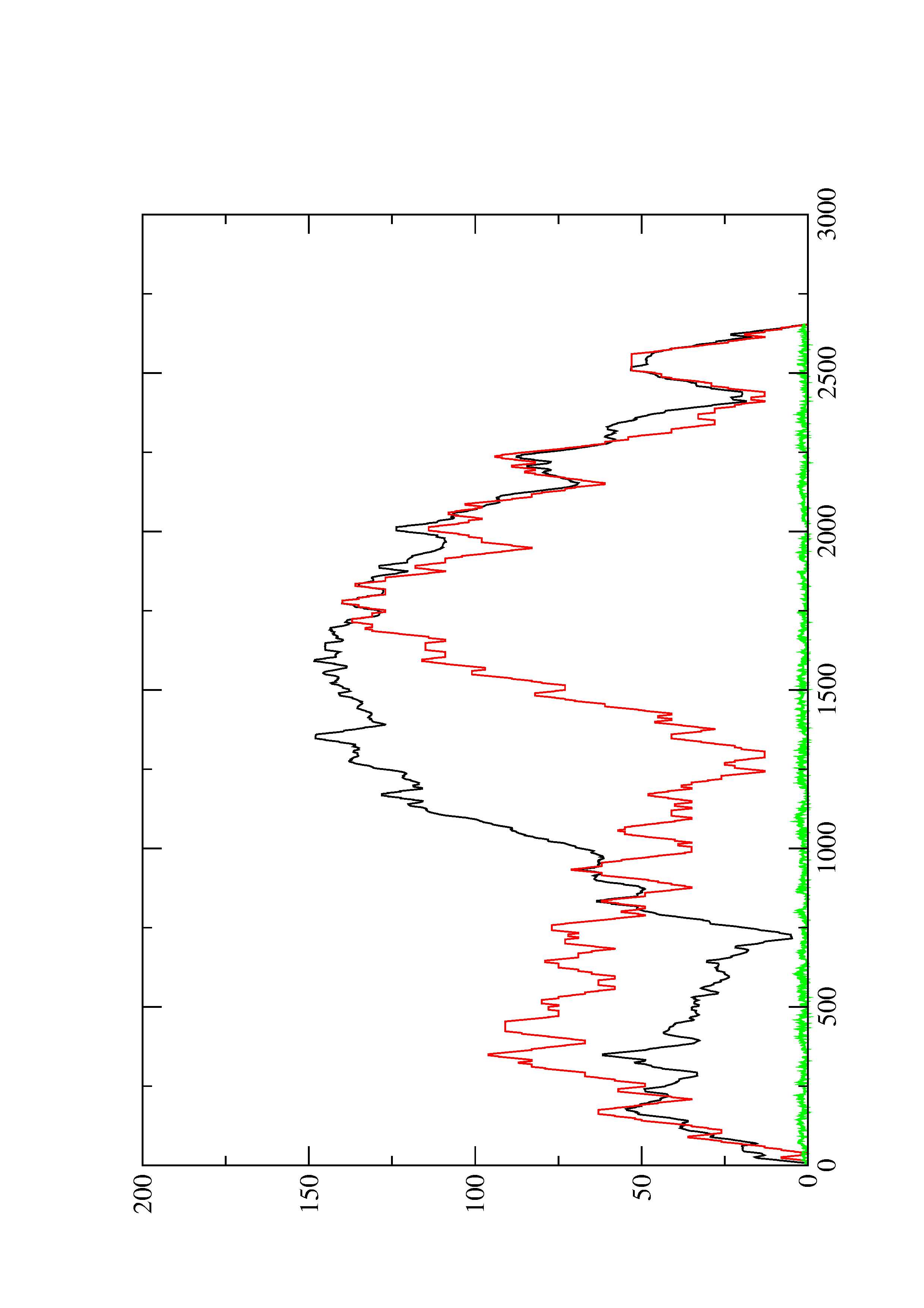
2. Here you find a mountain plot representation of the MFE structure, the thermodynamic ensemble of RNA structures, and the centroid structure. Additionally we present the positional entropy for each position. The resulting plot shows three curves, two mountain plots derived from the MFE structure (red) and the pairing probabilities (black) and a positional entropy curve (green). Well-defined regions are identified by low entropy.