lncRNA Structure Prediction
Species Name : Withania somnifera Sequence Id : GBHJ01014451.1
I. Result for minimum free energy prediction
The optimal secondary structure in dot-bracket notation with a minimum free energy of (-790.94) is given below.
1. Sequence-
UUAAUGAACAUCUAAGGGACUGAGUUCUUCCACUAAUUUCAAAUGCAACACUUUAUUUUU
GUUUUCGCAUUCCUUACUAAUUUUAUAUCCACAUUUUUUUAACUCUCUUAAGGACAUGCU
UUGCUAAAUUAUUUUUUGGGUCACACUCUGUGGUGUGUCACUUUACAAGUGAAAGCUAGA
AAGAUUGUUCCUCUCGCUAUCCUUUGCUCCUUCUGUGGUGUAUUUGGACGUUGGGGAACU
AUAUUAUAAUCCACAAGAUUAGUUUAAGGAAUUUGAUGCUUUAUAGUUUCCUGAUUUCUU
AAGGUUGCAGACUUUCCAUGGCCAAGAGUCUCUUUGAAAGACUUGAUUAGAAGUGAUGCA
UCAAACGAGUAGCAAAUCAAAGGAGUUUUGUUACUUGUACCUAUACUAUCUGUAGUGUUA
AGUGACUACAUACAUUAGUACUCCCUUUGAACCACUUUACUGUUCAUCCUUUUUAAAAAA
UAGAUUCCAUAAUACUUGUCAUUUUAUAAAAUACAUAAGUCAGGACAUAAACAAUGAAGA
ACGAAAUGUUUACAUAAACAAUGUCAGGACAAUUGAUUUCAGUGUUUGCAAUCAAAUCAA
UUUCUAAAAUCGAAGGAAAUAUUUAAUAUGAACAUGUUUUUAAGCUGUGAGUAGAAGGCU
UCUUAAUCUGGUUGGCAAUAAAUAAUCUCAUUUUGACCAACUCGUUCCCUUGAAGCAAUA
GUGUGUUGCCCUUUCUUAUUUCUCCUUCAUAACCCUUUCUCAUUGUGUUUGGUGUAUACU
GGGGCAGCUACAUUUGUUUGGUUUUCAAUUUCUAAAAGUAUUGAUGGGUUGAACAAGUGA
CCUGUAAUGAUUGGCGAGUAUGGUUGCAACAUUGUUUGAACAUCUAAGUGACUGAGUUGU
CCACCUCUUUGUAUGAUAAGAGAUUAGUUUAUAACUUGUACUAGAAAUGGCAUCUCCAUA
GGAGACUUUAAUCCCUCAGUCUGGAUAGAGUUUAUUUAUAUCAUGGAAGGAGACAUGAAG
AUACAUUAUAUUAAUGAGAGUUUUUAGUGUUACCUACAAUAGAGUUUUUAGUAUUACCUA
CAAUAGUGCAUGUGACACAUCUCUUUUUAAGGAUAACUAGAUUAUAAUCAAUAGUUUUGG
ACUCUCUGUUACUUCUUUUAUGGUUUAGAAGCUGCUCGUGGAGCAAGAAAUAAGUUUAAG
CACUCACUUGAACAACAAUAACUGAGCAUUGGUUCCUACUUCGGUUUUAGCUUCAACAAU
UGUGUAUGAUUAAUAACACACAGAAUCAGUCUUUAUAUGUGUUUUUCUGUGUCUAGUUGA
GUUCAGAAUGAUGUAGAAACAAGUAGUUUAAGAGCUGUCGCAAAGAAAACAAAUUAAGAA
CGUACUUGUAGCUUAUGCUAUGCCUAUUGCUAAGUAGCAAAGUAAAUGGAGAAAGUAUGG
CCGCUAUUUACCUGCCUAACGGUUCCCUUGUGUCUUUCUAUUCAUUAUGUAUAAGGUUGU
CUUAACAGAUCUAACUCUAAUCUUUAGUCGAAAAUAAAUAUCUCAACUUUCUCAGUCUAA
AUUACAAAUUAGGUUAUGGUUUUUAAUUUAGACUAACUGUGAAUAUGUAGUCUGCAUAAA
UAAUUUCCGGAAAUUUGAAACAUUUUUCCGUUUGGCUGCAUAUAUUAGAUGCAAAAUUAA
GAUGAUAUUCAAGUUGGUUUCAUUAUCAUGGUGACAAGAGUAUGUAAUGAUGAUAAGAGA
CCAAAUCUUAGUACCUCCAUGAUGUGGAUUAAACCCUUUGUUCAAUGAGAAUUAUCACGG
GUGACCCCAAGCUUCGGGAAACAAUAGCUUCAUCUUUGUGUUUAAUGUACUAACAUUAUC
UAUAAUGCUUAGAUGCAUUGAUAAGCAUCACAUCUAUGUAUUGAGCUGCUAUCUUUGUAU
UCACCAUUAUGGGAUCUAAGAUUUAUUGAUGCUCAUAAAUGCCUUUCUUGGUUGAGUGAC
AUCUUGGACACAUUUUAAGUGUACAUCGUGUUCUCGCCAGUUCCGUAAGUUGUGACAGCA
GAAAUAAAAGUGACUUGGUAGAAUGUGCCUCGUAUUUUGUUGUCAUACGAAAAAAAAAAA
AUUCAUCCUUGUGUAGUUCAGAAUUUGGUUGUUUGAGUUAACAUUAGAAUUUGUUAGUUC
UUUUGUGGCACAUAUGUUACAAUAAGAUAGUAAUUGGAUUAGGUCUUCUGUGCCAAAUCC
AUGAGAUGACAAUUUAAACAGCAUUUAAGUUUCAACCCAACUAACCUAAUGUGUUGUACU
CUGUUUCAAAAUAGAGUGAUAGUUUUGGGGCUAGCUAAUGUCAAUAUGCAUCUACUUGCC
UUACUUUUGAAUUCAUCUAGGAACUCUGGUUCUUGUUCCAAACUAUCAUGGCUUGCCAAG
GUUAGUAAUAGUUUUACGUGGCUUAACGAGACUUAAGCCAGGUGUCUACCACAGUCAUUU
GUUGUGCUGACCGUUGAAUGUCUGGAAUAGUAGACAAUGGUUCUUUGUUGUCCGAUAGUU
CGUUUGGUGACUAUUUGAGCUAACAUCAGAACCUUAGAUAUUUUUUCAUCUGAGACCUUG
CUCUCUGGGUUAGACUGCUCUUCCCUUUUAUUAAGGUUUGAGAUAAAUACACUGCGAUAA
UUUCAAGAUAGAUGCUUCUUUUUGUUUCUUUUCGAGACAAAUACAUGACUAUAUUUUAGG
UGUCUACCUCAGUCAUUGUUGUGCUGAUCGUUGAAUGUCUGGAAACAGUUAACAGUGGUU
CUUCGUUGCCUGAUAAUUCUGUUGGUGACUAUUCGAGCUACAUCAGAACCUCACAUUUAC
UUGACAGCCUGCUUUCUGUAUAGACCUUUUCUUCUCCUCUUUCACAAAUACAUAAGUAGA
UUUAGUCUGAAUGUCUACUCUUUUGCUUGCACAACUCACUGUAAGUUCCUUACUGAUAGA
UAGCAAUGCGGCACUUCUUUUAAAAAUGCAGUUCAGUAUUUUUUUUACCACAAGGCAUAA
AUAACUUAAUAUCCAUUUGCCAGGUUGAUUCCCUGCAACUUUGCGUAUUCUGUGAUGGAA
AGUGAGAUCUCCACGAACCUUUGUUGAUAAUUGUAUACUGAGACGUCGGGGAUGAUUACU
UAUUUGCCAGAAACUAUUUUUCCUAGUAAGAUGCUGAUGUGUCGAAAGAAGAAGUAAUUG
AUGCUGCUGUUUCUCUUAUAUUUUUAAAAAUCGAAAUGCCCAAUCAAUGUUAAAUUGUUG
UAAGGCCUUUUGAUUAUACAGAUGUAAUUAUGCUUUUCAUAAGUUUAUGUACAAUUGAGC
AUGAGAUACUUAUUUGUUGAGGCACUGUUUGUGGG
2. MFE structure-
...(((((((((.((((((.((((.............)))).(((((.((((((............((((((((((................................)))))).))))....(((((......)))))..((((((....))))))((((((...))))))..(((((((((...(((((((.((...(
((..(((.((......)).)))...)))))..))))))).......(((((.....))))).(((((((((((.((.(((....))).))..)))))))))))........)))))..))))...((((((.......)))))).....)))))).)))))...((((((((((((..........))))))))))))..
...(((((..((((((........)))))).....))))).)))))).))..........)))))))..........((((((...(((((....((.((..(((((.(((((((((.(((..(((((.....(((..((((.....)))).))).....)))))..))).((((((((..(((((...(((((.(((((
((.((((((((((((....................((((((((((((((((.((((((((((.......(((((((((..............))).)))))).........)))))...(((((((((((((....(((.((.....(((((..........)))))...)).)))....)))))))).)))))......
..........(((((((.(((((((..(((((((((.((...((((((.((........)).)))))).)))))))).)))......((((((.((((((((...........(((.(((((((..(((....((.(((((((.....((...((((...))))((((((.(((........))))))))).........
(((((........))))).....(((((...)))))((((.(((((.(((.....))).))))).)))).......)).....))))))).)))))..))))))).))).)))))))))).)))).))))))).)))))))..........)))))))))))))))))))))(((((...)))))........(((((((
......)))))))..........((((....))))...)))))))))))).......))))).)).)))))...)))))...)))))))).....)))))))))(((((((((...(((....)))...)))))))))(((((((((((..((..(((..........)))..))..)))).)))))))(((....))).
......(((((....))))).)))))..)).))..)))))...)))))).(((((..((((((..(((((((.................)))))))..((((((((.((.((((.........))))))..((((((.(((((((........(((((((((((.((((((.....)))))).)))))))))))......
..((((((((((...............((((((..((...))..))))))...))))))))))............((((((((..........((((((((.(((((((.((.(((......))).)).)))))))))))))))))))))))...(((((...)))))..........((((((((((......))..((
(....))).......((((((((.((((.((((((((....(((((((((((((((((....))))).)))).))))))))...(((((((((((..(((((((((((((((.(((((..((((..(((((((((.....((((((.((...))))))))........((((.(((.(((((..((.((((.......))
)).))..)))))))))))))))))))))...))))..))))).))).((((((((((((((((((((((((((((.........)))))).........((((((.((((.((.....)).))...)).....)))))).(((..((((((....))))))..))))))))))..(((((.........)))))((((((
.(((.......))).))))))..((((((..((((((((((((((((.(((...........))).))).))))))(((((((((....))))))))).......((((((.((...((((......))))...)).))))))..))))))))))))).(((((..........)))))(((((((..((((((....))
))))...)))))))(((.((((((((......)))))))).)))))))))).))))))))...((((((..(((((..(((((((((.((((((((..((((((.(((((((.(((((((........))))))).))..)))))..))))))((((((........((((.((((((((......(((..(((....))
).)))......)))))))).)))).........((((((((....((.(((((((((...((((((((.....))))))))................))))))))).)).....))))))))((((.(((.((((((((((....)))......(((((((...((((((.((((....)))))))))).....))))))
)...........)))))))..)))))))((((((..((((((((........)))............))))).)))))).....)))))).)))))))).))).(((((((........)))))))..........)))))).))))))))))).............))))))))))))..............((((...
...........(((((((((.((((((........)))))).)))........))))))((((((.(((((((((...(((.((..((....))..)).))).))).))))))..))))))...))))....((((.......)))).))))).)))))).......)))))......))).)))).)))))))).....
........((((((((.((((((.((((.....)))))))...))).)))))))).......((((..((.((((.....)))).))..))))))))))))))))))).)))))))))))))))))))).)))))
You can download the minimum free energy (MFE) structure in
II. Results for thermodynamic ensemble prediction
1. The free energy of the thermodynamic ensemble is [-849.45] kcal/mol.
2. The frequency of mfe structure in ensemble 5.86631e-42.
3. The ensemble diversity 1083.74.
4. You may look at the dot plot containing the base pair probabilities [below]

III. Graphical output
1. A note on base-pairing probabilities:--
The structure below is colored by base-pairing probabilities. For unpaired regions the color denotes the probability of being unpaired. The reliability information of RNA secondary structure
plot in the form of color annotation, Positional entropy ranging from red (low entropy, well-defined) via green to blue and
violet (high entropy, ill-defined).
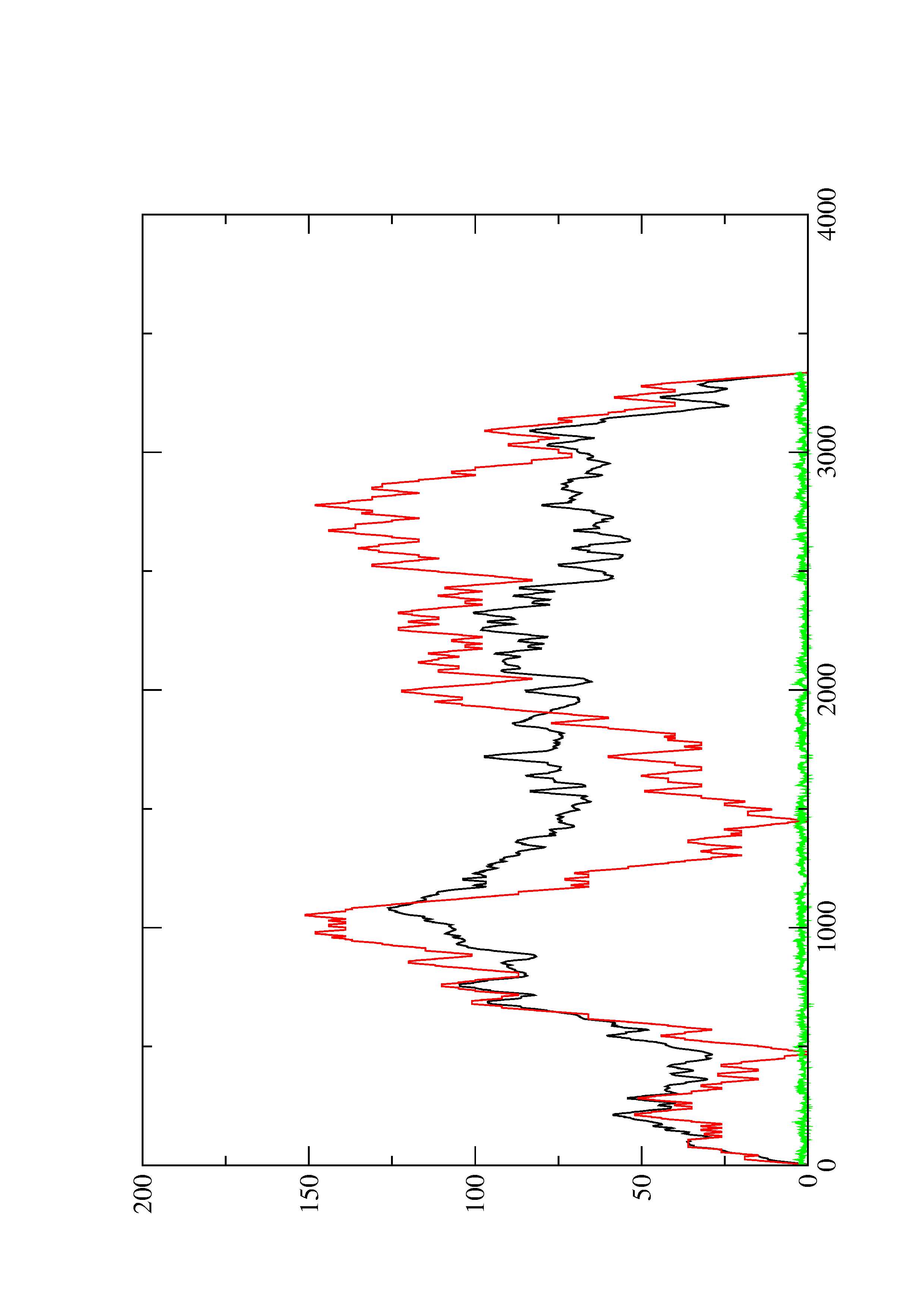
2. Here you find a mountain plot representation of the MFE structure, the thermodynamic ensemble of RNA structures, and the centroid structure. Additionally we present the positional entropy for each position. The resulting plot shows three curves, two mountain plots derived from the MFE structure (red) and the pairing probabilities (black) and a positional entropy curve (green). Well-defined regions are identified by low entropy.