lncRNA Structure Prediction
Species Name : Withania somnifera Sequence Id : GBHJ01014825.1
I. Result for minimum free energy prediction
The optimal secondary structure in dot-bracket notation with a minimum free energy of (-486.80) is given below.
1. Sequence-
CUCCAUUUUAUAUUUAUUUGAUAAAAAUUCAGAAAUUACAACCAUUGGGCUGCACUAGAU
UCCAUACAAGUAAAAUAAAAAUGCAUACAUAAGUAAAUACAUAUGGAAAAGAGGAGAAAC
AAGCAAAAUCUUUAAUUGGAUCGAGUCCAAUUCUUCCAACCUUGGGUCAUAUUCAUGAAC
UCGUGCAAGGAAUUCAAGGUUAUACAUGCAAAGGAAGAAAAGAACUUAGAGACUAUCGAU
AAUAAAUAAUGUAAACAAGUCAAGGUAACAUGGCCAUGAAGUUAUAUUGCUCCAACCCCA
AUUAUAUAAAAUUCGAAAUGGAGAACAUAAAGAUAAGCGGCCCUAUAGAACCAAUCAAGA
GAAAUGGGCUAAUAAAACUUUAAGAAGGGCCUAAUUAAUUCAAAUACACCGUGGACCUAC
AUAGGCAUGGUCCAUCCCAAUUGCAUAUAUCUAAGGUAAACUUACUAAUAUUUAACUGAA
ACUUAAUUAUCUAAGUGAAAACAUCCAUUAUACAAGAAUAUCAUGAAGACUUUGAGUUAA
UCAAGUAUGUGUGAUAGAAUUGGGUUGAGAUAUAGGAGGAAGUAAAUAAAUAGAAAAUUU
CACAGAAGAAAGAAACGACAUGAACUGUACAUAAAGGAAGAGAAACAAAAAGCAAGUGUA
CAUGGAUAAUAAGACCUAAGCUCACUUAUAUAAGGAAAUUGGCAGCAUGCUUCACAGAUA
ACUCAAUGCACAUGAAAAGAAACUUUAGAAAAGCAUCUAGUAGAAUAUGAUAUUAACUUA
GCACAACGGGGAAGCUAGAUAUAUGACAGUUUCACAGAAAGAGGUAAUACCGCUAUAGUU
AUGAAGAACAUGGAUUAUUGGGCUACAGGUUCAACAAAUAGAGAAAGGAAAUGGGAGAAC
AAGUGCAUAAUAUAACUUAACCCAAUAUCAUAGAAAGGGAAAAGGGAGGGGUGACAGGUU
UAUGAUAUGUACUAAAACAGGCAAAGGUAGAUAGGCAACAAGCAUGCAUAAUAAGACAUG
AGUAUAAUAACUUAGUAAAGAAUGAAACAUGUUCAUAAUAUAUCACUUAAACCUGGCUUA
AAAGAGAGGUGAAACAAACACCAACUUAUAGAAGACCACACGAACUCAGUAUAAAAGAGU
AAAAAUAAGUAGCACCUUUAAAAAUAUACAUAAUGAGUUUAAUAUAAGGUGAGGAAGGAA
ACAAGUAGUAUCUUAGAUGAUUUUGGAAAUGAAACAACAUGCAGGUAUUGAAGGAAAGGG
GGCACUUUAUGAUGCAUAGAUUUAUAUUUCAGGGAAAAGUAAGCAGUCAAAGUGAAAAGC
AACCAACCAAGUAUUUAUGAACAUGAAGCAACAAAAAGAAGCAGUUAAAUAAAGAGAGUU
GACAGGUUAGUACAGGUUUUACAGCAACAAAGUGAUUACAUGGGAGCAACCUGAAGCAGU
CAUAAGAAGAUAAAACUAGAAAAAUAGCAAGUAACCUCAGUCAAGCAAGUAGCACCUACU
CACAUAUCAUAGACAGAAUUAGAAUACAGGACAACAGACUCCUUCUUUAACCAUUUGAUC
AAGUUUUACAUAUGGAACCAUUUAAGAAAAGUGCAUGCUCAAGUUUAUGUACAGCUUAGA
AAAGAACAGAUCAAGCAAGCAUUAUGAUAGACUCAGUGGAACGGAGUUCACAGAACAUGU
UAGACUAACAUUAGAAGCCCAAUGCAUUUAUCCUAAACAUGUUAUAUAAUUUAACUCAUG
UGAGCAGGUAUUUGAACUCAAUCAAGCACUAGGCAACAGGCACACUACGUAGGACCAAAU
GAAUUUUUCUCAAAUAGUUAACUGGCCAACAUAAGAGGAUUUAUGAAUUGAAAACUCAAG
AACUAAAUAACAUACUCAACACAAGGCAAAUGAAUAACAUAAACACCACUAGAACAGGUA
GGUUAUGCGCACAGUAAUAACAGUAAGUAUUGUCAUCAUUCGGUCCAAGUUGAGGCAGUU
UAGGUAGAAUAAUCUCAUUAUACUGAGUUGUGGAAAUCAUAUUAAUGAUAACAUAGGAAA
ACUUUCAAUAUACUUAAUUAGUUUAUGAGAGUGAUCUUGAAACUAGCUUUUCAGUUAAAG
AAAGGCAUGUAGACAAGUUGCACUUACAAAUCAGCUCAUACCAUAAAAUGAAUAUCAUUU
UAGCCCUAGUUAACACGCUCACACAUUCAAGAAAAGAUCACACCCAGUAAACAAUGAGUA
GAUCAAGGAAGUGUGAUAUUAUUAUCCCUAUUCAAUGUUAAGGGGAAGAGAAAGAGUAGG
CAUGAUAUCAACUACCUUAACUAAGCAAGGGCAUAGAACAGUAUAUGAUCAGCAUGUAGG
GCAAAAUAGAAACAAGCAAUAUACUAGCUAUACCCAUAAAUAGUAACAUGUAAGGUUGGG
ACAUAGUCAUACACUCAAAUGCAAAAUGUUAAAACUAUGGGAGAAAUAGACAAAUUUGUG
UAAGAAGAAAAGGCUCUCAUAGGCAAUAGACAAAUAAUUUAGAGAAAUAGGAAUGAAGCA
ACUACAAAUAAUGCAUGCACAUGCAUGAAGCAAUAGGUAUCAUAUAUUCAGAAUGUCUGG
AUCUAAAAUUCAGGACAUGAUGACAUCUACAAUCCUCUUGUAGACAAGCCAAACCCUAUA
GAGCUACGCGCAAUGGGU
2. MFE structure-
..........((.((((((((.(((((((((...........(((((((((...(((..(((((((...(((........(((....))).......))).)))))))..(((((.............))))).(((((((...)))))))((((((((((((((((......((((....))))......)))))))))
)...........))))))..........(((.(((((.(((((...((((((((.....(((.(((......))).)))..))))))))(((((.....((((......)))).....)))))....(((((......(((((......................))))).......))))).....((((((((.((((
((........(((((((...........)))))))...(((((((((((((((.((((....))))...............((((.(((((((.((((((((.(((.((((((((...(((((((...((((.((.....))))))...)))))))...)))........)))))..))).))..............)))
)))......................(((((((.......................))))))))))))))))))..((((..((.(((....)))))..))))......(((((.......(((..((((......((((...))))......))))..)))...............(((........)))))))))))))
))))).)))))........((.(((...))).))...(((......))).))))))))))))))...(((((......(((((.....((((((......((((..........))))..(((.............)))..(((((((((((.....(((((((((((((.........(((..((((..((..((....
(((.(((........((.(((((......)))))))......((...((((((((((((((.((((((.....((((((....(((.((((.......))))..)))....))).)))...((((((((((((((((.((.............)))))).....)))))....)))))))......))))))........
.....((((((((...(((...(((.........))))))..))))))))..((...((..((..(((((..((.((...(((((.((((...)))).)))))..))))..)))))..))..))..))..))).)))))))))))...))..........)))))).......((..(((..((((((.((.((.((.((
((.........))))..)).))...)))))))).)))..)).........................))..)).))))..))).....(((((...))))).))))))))))))).......................)))))))))))..)))))).......)))))......)))))...........(((((((...
.....))))))).((((.((..........))))))....)))))))))).))).(((((......))))).....((((((...))))))))).)))))))))......(((((.(((((...............)))))..((.(((.(((((......))))).)))..))...((.....))...))))).....)
)))))))).)))))))).))..((((....(((((((((((......((((....))))...((((((......((((((....(((.(((((....((((...((((((......))).)))))))...((((((...........))))))..)))))..)))...))))))...))))))(((((......((((((
(.(((((((.((((.(((((....)))))....(((((..(((((((.((.(((.....))).)))))))))..)))))......(((((((.......))))))).(((((((..(((.((((((((.....((((......((((((((...))))))))(((((.(((........................((((.
.((.((........)).)).))))((((.(((.(((((((((....(((((((....................))))))).)))))))))))).)))).....))).))))).........((((((.....))))))))))...................(((((..(((....)))..)))))...))))))..)).)
))...))).))))(((((((......((((....(((((((((...((.(((....))).))...........)))))))))......))))....)))).)))........((((.((...........((((((....))))))..........)).))))...((((((...)))))))))).)))))))...))))
)))..))))))))))))))))......)))).((((......((...)).....))))
You can download the minimum free energy (MFE) structure in
II. Results for thermodynamic ensemble prediction
1. The free energy of the thermodynamic ensemble is [-536.71] kcal/mol.
2. The frequency of mfe structure in ensemble 6.74112e-36.
3. The ensemble diversity 602.41.
4. You may look at the dot plot containing the base pair probabilities [below]
III. Graphical output
1. A note on base-pairing probabilities:--
The structure below is colored by base-pairing probabilities. For unpaired regions the color denotes the probability of being unpaired. The reliability information of RNA secondary structure
plot in the form of color annotation, Positional entropy ranging from red (low entropy, well-defined) via green to blue and
violet (high entropy, ill-defined).
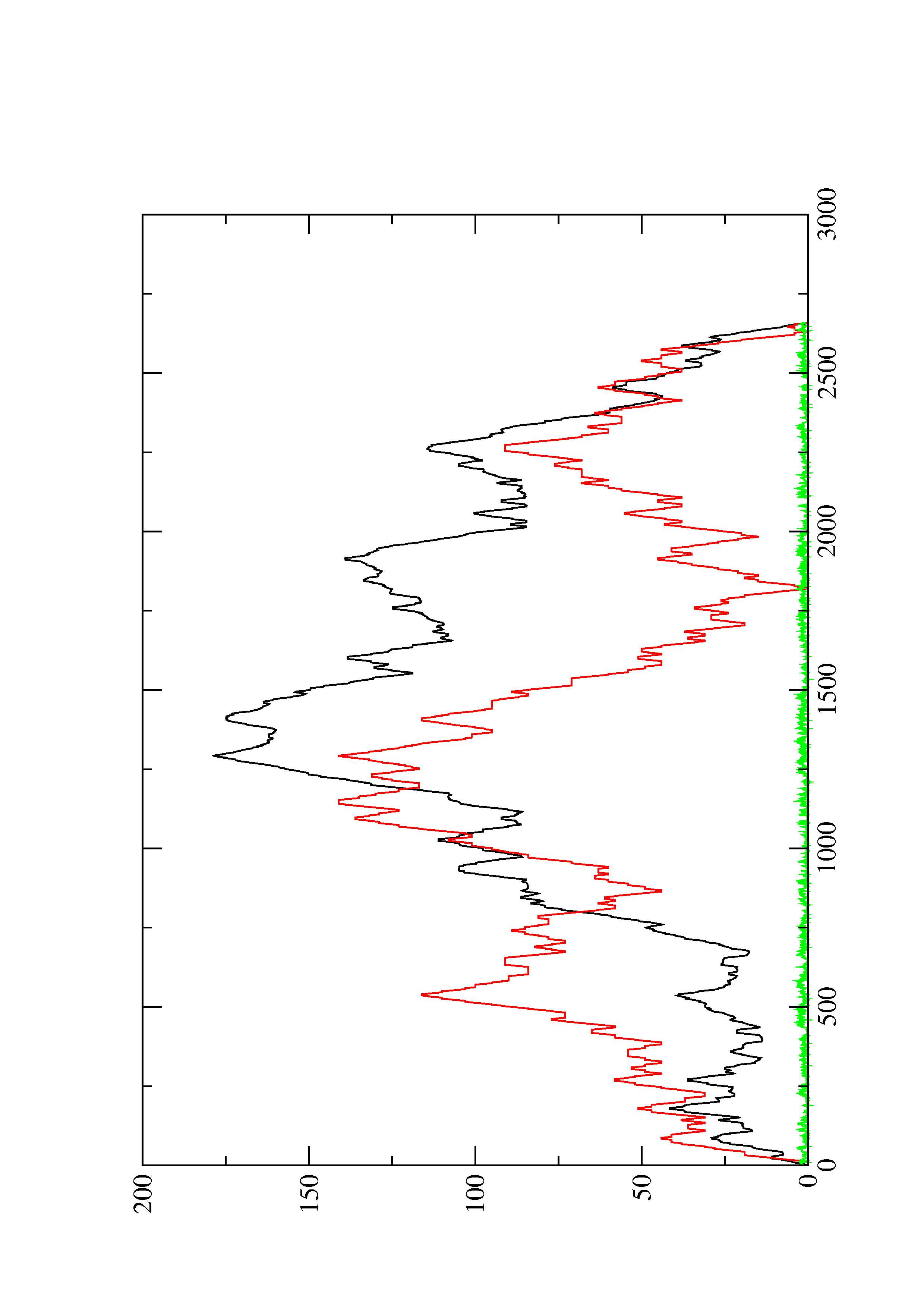
2. Here you find a mountain plot representation of the MFE structure, the thermodynamic ensemble of RNA structures, and the centroid structure. Additionally we present the positional entropy for each position. The resulting plot shows three curves, two mountain plots derived from the MFE structure (red) and the pairing probabilities (black) and a positional entropy curve (green). Well-defined regions are identified by low entropy.