lncRNA Structure Prediction
Species Name : Withania somnifera Sequence Id : GBHJ01015178.1
I. Result for minimum free energy prediction
The optimal secondary structure in dot-bracket notation with a minimum free energy of (-435.20) is given below.
1. Sequence-
UUUUUUUUUCAACAAAAAGAGAAAUAGGUGCAAUAUCAAAAUUCUAACUACAAGCCACUC
AAAAAUUAGUAGCCUCUAAAUAUGAGUAAUACAAAAUCAAAAUAAAUACAUAAUCUUAAU
AUCUCAAAAAUCCUCGAAGAAGAUCCAAGACAAAAACAUAAAAGAGUUUGGGUAUCUGCU
GGAUGAACAAGGCAGCUACCUUAACUCUGAUAAACUCGAAUCACAAGCCUGAUCGGGAAG
CACAAAUCAAGUUGAACUGAGUUGUGCACCUACACAAGUGUAGAAAGCAAGGGAGCAAGU
ACAAAAUAAACAAUGGGUAUCUAGCAAAUAUCCCUGGCAUGGCACUAAAGCAAAGUGGUA
AAGAGGUGACGAGUCACCGACUACCGCAACCAUACCAACCACAAUCCUCCAUAUACCGAA
CUGCAAAUCAGACCAUAAUCAACAGUUCAUAGUAAAAAAUAAACAAUAAAUACAAGAUGU
AAGGGUGAUAAAGGUAAUUAACACCUCAGCAGAAACCAAAAGAAAAUCAAAAUCAGGAAA
CCAGAGUAAUAAUCUACAACUGAAACCAUAUCCAAAAUAAGAGAGUAUCAACAAGUCACC
GCCAGCCAACCCGAAUAAUGCAUCAACACCAAGUAAAACCAAUCCAAUUCAUAUCAAAUG
UUCAAUUUCACAACUCACGAGAUGAAUCUCAUUGAUUUUGCUAUGGCCUCAUAGGCAGAG
UCCAUAAAACCUCAAUGAGCUCCCAAUUCACAAUGAAAAACGGACUCAUUAGCUGGAUAC
UAAUGAGAAUCAACCAAGCUAAAUAGAGGAACGCAACUUAAGUCUAAUACACAUGUCAGU
UCCUACCUCACAAGUCUAACCGCAGUAUGAAAUGUACACCCGAUGCAACUGAGAAAUGGG
CUUGUAGGAUAAAUUCUGAAAUGCCUAGACCACACAUAAGUCUACCUGCGAUAGGAAGAG
UAAAUCUAACGCUGUCAAAAAAUAGACUUGUAGGAUAGACUUUAAAAUGUAUAGACCAAC
AUCAAUACUAUACUCAACCAAUAAAGUCACUUUUCAUUAUUUUCUAAACAUUUUAGGAGG
CUUUCUAUUUUAAUCUCAUAGUAGUACAAUGCUCCUCAACUCAUCGGGGUUAUAAUCAAG
UCCUAAAAUUUAAUCAUAAUCAACCAUCUCGAGCUCGUAAUAUCAUCCUAAUAACAAAAU
AAGUGAGGCAAAAUAAAAAGUGAAAAAGUACUCCAAAACAGACAUGCUCACGAAUUAUGA
AAUCAAUAAAGUAAACAAGUCCAUGUAUGUGCAGUGCAAUGCAAUACAAUUUCAUGAAAU
GUAACAUAUCCUGGUACCUCACAUCACAUAUCUUUACCUUUACCAUACUGAAAUCACACA
UCGUCCCUGGUAUCUCAGAUCAUAAACAAUAACCUUACCACACCGGAAUCAUAAAUCAGC
CAUGAUUUUUCAUAUCACAUAUCUUUACAUUUACCACGCCGAAAUCAUACAUCAGUCAUA
AGUCUUUCAUAUCACAUAUCUUUACUUCUAUCACGCCGAAAUCAUAUAUCAGCCAUAGUC
UUACACAUAACAUUCCUUUUUGACUUUCUCAUUCAUCAUUUACACACCAUAUACCUCAAA
AUCAUGUUCCUUAACCAUAUCCAAAUGCUUACAGCAUGAAAAAUAUCAUAUGCAAUUCAC
GAAAAAGAGGCAAUAUCACUCAAGACAUCAAUAAGGCGACAUAUGGGCACCACAACAAAG
AGGAAUUAUAUAAAUCUCAACCAAAGCCACUCAAGAGCAUGAAAUCAAAUAAGGGAGUAA
GCAAGUGUAAUUAAUAAUAAUUCGGCGUCUCAUGGCAACGUAGCUACUUUAAUCCAUAGG
CUCAUAUAUCUAAUUUUUUUAACUUUGUUAUAGACCCCGUAUUCUAAACUCUAGUAUCAG
AGUGGCUAAACUCUAGUAUCAGAGUGCUCAUUACUCCCCAUUUCAUAAACAAUUUAUAAA
AUCCCUAGGAUACUGUGUAAGAGAGGCAAAGGCGUCUACUUGCCUUAGAAAGCAAAGAAA
UAAUUAAACGCGAGCUUUUCCCUUCCGUUGAGCUUCCAAAUCCGCAAAAUCUAUUCAAAU
AUGAACUAAGUAUGAGAAAAGAUAAACAUUGAUACCCAUAUUGAUAUUUUUAAAAUCGGG
UCCAAAGCUCCUCGAAGAUUCUUGGAUCAAAAUGCUUGUUAUGUCCUCAAAGAGAACACC
UUAUGCAAUAAAGAAUACCUAAUACCGACCAAACAUAAUACGAAUUUAUUCUACCUAAUA
GAAACAACCUAGAUCUAACAAGUCACUAGCAACAACAACAUGAUUUUCUAAGGUUACUUC
AUUAGUAAAGACUUUGAAAAUGAUGCACAAACAAACAAGUCCAAUUUUAGCAUGCUUUGG
CCCAAACAGGAUUCUACUCAUGAUGAAGUAUACAAGCUUUAUUAAGUAGUCCACAGUAAA
CAUCCAAACCAG
2. MFE structure-
....((((((........))))))...(((((.(((((((..(((....((((((.(((((....((((......))))...)))))......................................................(((((((((............((((((.(((((.(((((..........))))).))))
).))))))......(((((.(((......))))))))..((((((.(((........))).))))))..(((((....))))).......(((((((.((((.............((((((((((.........((((.((((...........((((((....(((((....)))))...)))))).............
.................((((((...................))))))......................((((...))))..((((((..((((.......))))..........................((((......(((.......)))....)))).................((.....)).....))))))
)))))))).......................(((((((.((((...((((((.((....(((.........)))....)).))))))...)))).)))))))((((.(((.......))).))))........((((((.(((...((((...))))....))))))))).)))))))))).((((((.......(((((
......((((((...((.................))..)))))).(((.(((((((((....((((.((...((......))...)))))).....)))))))))))).....((((((((((..((((......(((((((((.....(((((..(((.....)))....)))))......)))))))))......(((
((((....((((((.............))))))........)))))))..............))))..)))))))))))))))..........))))))....))))..))))))).........((((((((.((.((((......)))).)).))))))..)).....((((((((......................
..((((((..........((((......))))......(((..(((...(((..(((((((((....................(((((.((((......))))))))))))))))))..)))..)))...)))...))))))........(((((..((((.....(((((.........(((....(((((....((((
.................((.....))......((((((....))))))............)))).......)))))...))).........))))).....(((((((.((.((((((((((............(((.....(((((.((.(((.(((((((............(((((((((.....))..........
.......(((((.........(((((((...(((.(((......))).))).)))))))........)))))...........)))))))...........))))).....)).))))).))))))))............(((.(((.((((((((((................))).(((((((.......((((((((
...(((............(((.(((.((((.(((((((.((((((.............))))...................)).))))))))))).))).)))....(((((.((((.(((((......))))).)))).))))))))..)))))))).))))))).....))))))).))).))))))))).)))))).
))))))).((((((......))))))..)))).)))))...........))))))))....(((.((..((((((....(((((....((((.(((..((((.......))))..))).))))....(((((((....)))))))...........)))))...))))))..)))))..))))))))).....)))))).
.(((.(((...))).))).............((((((.............................))))))..(((((.(((..((((((((......((((((...............))))))))).)))))..))))))))...))).)))))...)).)))))........................((.(((((
.....((.(((..(((((..(((((((((......)))))))))))))))))...))......))))).)).
You can download the minimum free energy (MFE) structure in
II. Results for thermodynamic ensemble prediction
1. The free energy of the thermodynamic ensemble is [-474.16] kcal/mol.
2. The frequency of mfe structure in ensemble 3.53246e-28.
3. The ensemble diversity 443.62.
4. You may look at the dot plot containing the base pair probabilities [below]

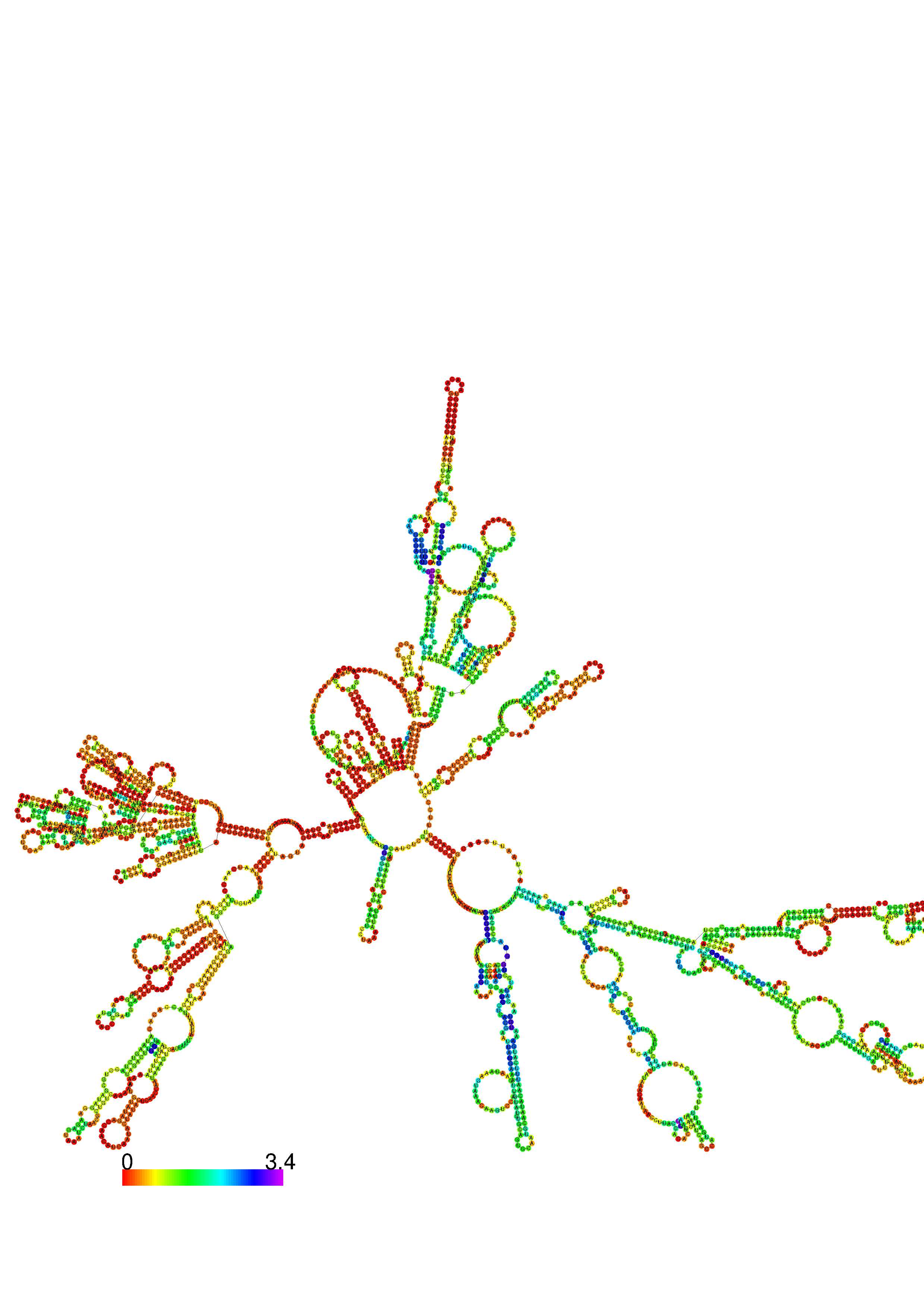
III. Graphical output
1. A note on base-pairing probabilities:--
The structure below is colored by base-pairing probabilities. For unpaired regions the color denotes the probability of being unpaired. The reliability information of RNA secondary structure
plot in the form of color annotation, Positional entropy ranging from red (low entropy, well-defined) via green to blue and
violet (high entropy, ill-defined).
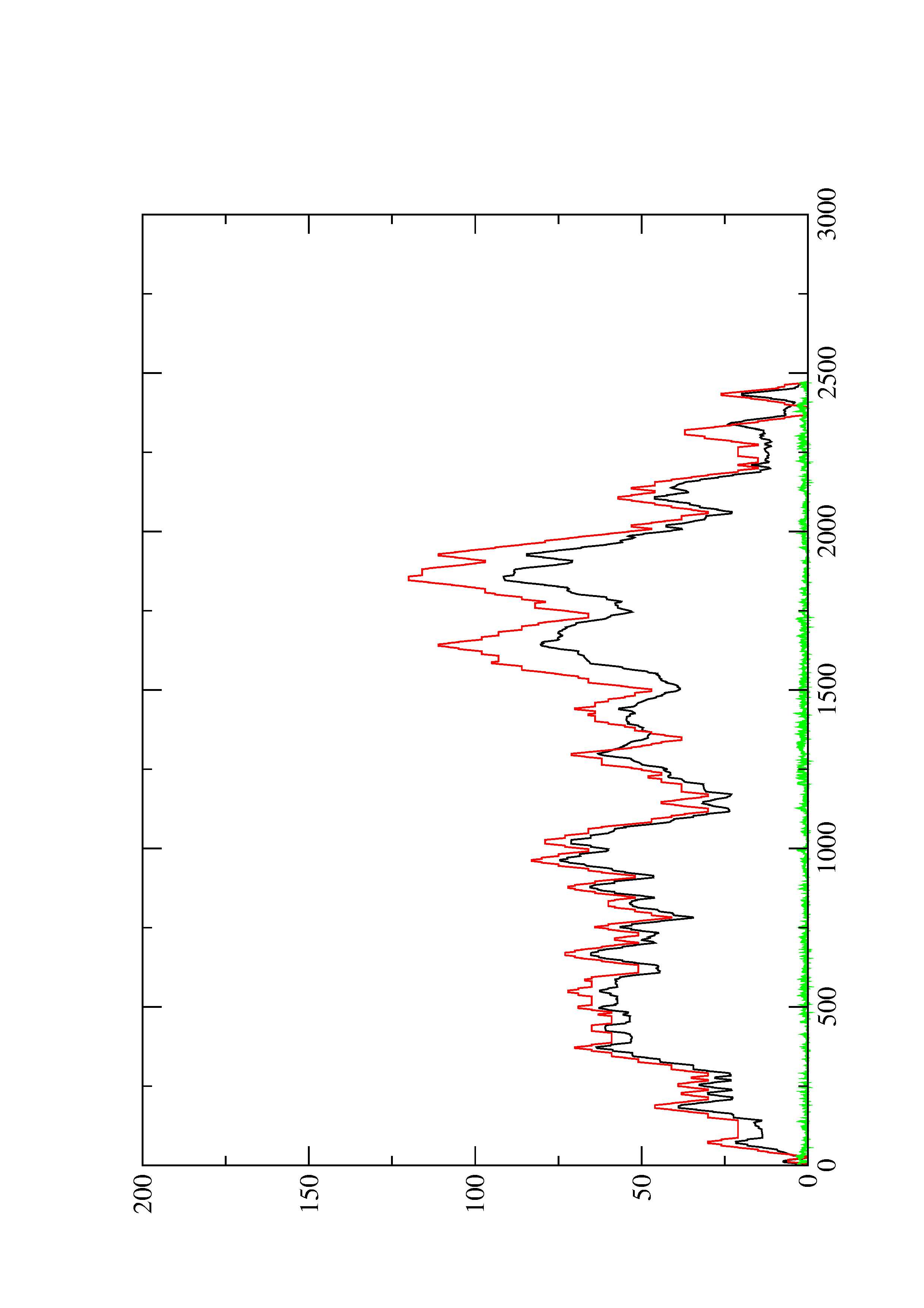
2. Here you find a mountain plot representation of the MFE structure, the thermodynamic ensemble of RNA structures, and the centroid structure. Additionally we present the positional entropy for each position. The resulting plot shows three curves, two mountain plots derived from the MFE structure (red) and the pairing probabilities (black) and a positional entropy curve (green). Well-defined regions are identified by low entropy.