lncRNA Structure Prediction
Species Name : Withania somnifera Sequence Id : GBHJ01015336.1
I. Result for minimum free energy prediction
The optimal secondary structure in dot-bracket notation with a minimum free energy of (-494.89) is given below.
1. Sequence-
AUCUUAUUAAUGUCAAAAAUCAGAGAUCUAAAGCUCCUGUUCCCGAAACUUGAUUGCAUA
CAGAUAUACCAAAUGCAACUAAAUAAAUAACACAGAAAAAAAAAACUUUACAAAAUAAGG
AGAGAAAAGGAGGCAAACUUAUAUAUGGGCCAUAGUCCAUGCAAAGAAAGCCUAGGGUAA
CACAUGCAAAGAAAGAAAAAGGAAUUAGGAAUCGAUAAUGCUAAAUAAUAUAAGUAAAUA
GAUAAAUGAAACAAAUAAAGAUAAAUGCAACCAUGAAAUCUUAUACUGGGCUAGGCCAAA
AUAUACAUGGUUCGGAAAAUAUAACAAGGGACGGUCCGGAAGAAACAGACUUAGUUAAUU
AUAGAAAACUGGGCUAAAAAAGCUAAAAACAUGACCAAUUAAGCUAAAACCAAACACCAU
GGGCCAAUCUACUUGCACAGCUGCAGUCCGGCCCAAUUUGAUUAAUUCAAGACAAGUAAA
CAAGUAAAUAGCCCAUUUAACAUAUAUUAUUUAUCAUGUAGGGAAUAUAGAUACUAAUCU
UCAAAUUUUAGGUCUAGGUAUCUGGGUGAUAUACACACAUUAUAUACUUAAAAUAUUAUG
GAAAGUUAAAACAAGACAAAUAUAGGCACUUAGUCUUCACAUAAUCUAGGGAUUAACCUC
AUGAGCUAUUAUUAGUCAACCAAAUAGUUCACAAAGCAGGCAUAUGCAUGAACGCAUGUA
CAACACUAAGUACUAACAACAUGUUAAGACCAGAUAAUCAGGCAAUAUGAAUAUCACUAG
AAGGAAGGCAGAUCAUAGUGACAACAUUUAGACUCAGCACUUACCUAGUUUUUGUCUUCA
AUUUAGGCAGGUAGUUUAGGUCCUUAUACCAAGAUAAAAGGUUAUACAUAGGAGAUUACU
CAUUUUAGUUAGUUAACUCAGCCAAAUAAGCAUGCAACAGACUAGUAGGUGAAUAGAGAC
ACAUAAGAAAAUGACAUAUAGAACUAACACUUAUUAAAGCAUGCAUGACACCUCAAGGUU
UCAAUAAAACAUAAUUUUGCAUGUAAUCAGCAAUCCUAGUCUAACAAGCCCUAAAGACUA
CACAUUAAACACUCAAUUUCCAAUUUUAGUUUCAAGUUAAAACUCAACAAGACUUAACUA
GAAACUAACUUAUAUAUACUCCUCAACAAGUCAUCGAACAUAUAUAAACUAUUUACUACA
UUACCAGCACAAUUAUUCAGAAUGACAUACAAGUCUAAGGGUAUAGAAAAAAUGAAACAG
AGCCAGGACUCCAAAUAAAGAGCAUUUUAACCUACUAAGUAGAGAUAAAGGCACAUACCU
AGUGAAUGAAUGAUUCAAACUUUAAUACACACAGUAGAUAAUAGCAUCUCCAGUUCAAUU
AUCAAGUUGGUAUUUUUCAACAGUCAACCAGGACACACCCAGACAUGCUUUUAAUCAGUC
AUUUAGAUAAAAAUACCAUGAAUAAAUCCUUGGGACAAUUUACUAAAAAUUAAAAGCAAU
GUUGACAUCAAUAAUACUAAAAAAUGGAAAGUAGGAUAAAGAAUUAGUAGAGAGGCUUUU
UGAGUUUGAAAGUCAUAUAAAAAAAUGGAGAGGAAAUUUAAGUGUCCUCCAUCCCUCAAC
CCUCUGUGAAUUCAUACCUUUGGAUUCCUUGGAGAUGAGUUGGACCACACACCAACUCAU
ACCCCUCGCCUUAGGAACUCUUGCAAGACAUAAGACACUGGGAUUGACAAAACAAGCAAU
GGAUUCCCAGAAUUAGCCUUACAAGUCAACUCCCUUUCUUUGCACUCAUUAAAGUCCUUG
AAUAGUAUAGGCAAAGGAGGAAGGAGGUAAGGUCUAAACUUUCACCUUAAGUUAUUUUGA
AACUAAAAUCAAGACUCAUGGAUGAUCAUUUUUAGCACAUAAGGCUCACAUAUGGACAAU
GUUGAAAAUCAGAAUAUGGGGCGAGUUGAGGGGUCUUUUCUUUAUGACUUUGGGGUUGUC
AAAGUGACUAUAUGAAAGUCACCUUUAAAAACAGGGAUCACAUGGCACUAUGAUGGCCUC
CCAUACCCUACUAAUCAAAUAGGAAACUUGAUUAUACCUAAGCCAUUUAGUUUUGAAAAG
AAAGUUUAUAAAAUAUUCUAGGGCUAAAGGUAUGAAUCAAAAAUUAUUUUUGACAAGACU
AGAUCAUAGACAGCCAGUAGUCAAAUCACAUAACCCCACAUAGAAAAAUAAGUUGAUUGA
CUAACUUGGUCAACAAGAAAGACUAGAAGUGAACAUAAGCUAACAAAAUGAAUUGACACA
UAGACUUUAGACCAAGAACACCACAACAAUUAACAGCACAACAAUUAACAGCUAACAGAC
AAGUUCAAAUAACUAACACUGACAAGUAUGUAAGAGGUGUUCAUAGACAGAACUAUUCAA
CACAACACAUAGUUAGUUCAGAUAUCGUACUAAUUUAAAAUGACUUGUUUUAAUCCUGCA
CAUAAAUUCUACUUACACAGACCAGAGAAUAAAAUUAAAUCCAAAGACUACCUCCUAGAU
CCCUCAUAAACUAGGGUCAAGGAUAAAUCAAAGAAGGACAUACUGACAAACAGGUUUAAU
CUACCCUGAUGAUAAGAAAAAGAAACCAAC
2. MFE structure-
.((((.((..(((((....((((((((...(((((((((((((.....(((..((((((.......((((...................................(((((.....)))))..........((((...(((...(((((((....)))))))..)))...))))..))))....))))))...))).....
)))).))))).......(((.(((((((((((.((......................(((((..(((....)))...)))))....(((((((..........(((.((.((((((..................))))))......((.(((((((..........))))))))).....((((................
.))))..............((((((.....(((.(((....))))))..))))))..((((.....))))..)).)))..........)))))))....)).))))))))))).)))(((((.....((((((((.((((.........))))...)))))))).(((.....))).(((.((((.......)))).)))
....(((((((((((.((.....((((.....))))...........(((......)))..(((((((((............)))))))))..........((((((......)))))).........((((((((((...)))))......((((.(((.......))).))))......((((((((.((((..((((
....))))..)).)).)).))).)))(((((.(((..((((((((......(((((((((((((((((((.........))).....))))).))))..(((((((((((((((((.(((((.......((((((.......(((((((((..(((...........................))).)))))))))..))
)))).(((.(((....))).))).............((((.((....))))))...((((((............)))))).........................((((((((.((((((..((.....))..)))))).)))))))).........((((((((((..((((((.........................
...................)).)))).....(((((((((((((.(((........((((((....((((.((((((((((((.((((...(((((((..........(((......)))..(((((.....)))))........(((.....)))(((((((((.......)))..))))))..(((((......))))
).((((((..((......)).....((((((((((......(((((.((((....(((.((.....))..))).....))))))))).))))))))))..))))))....((..((((...........))))..))......))))))))))))))))))..)))))..))))............((((((((.((...
.)).)))))..))).......))))))...))))))))))))))))....((..(((((((((........)))))))))..)).((((((((.....(((((.......)))))..((((((....((..........))...))))))..((((((((((.......(((((((((((((((((..((((.....)))
)..)))....))))))))))..)))))))))).))))....(((((...((((...((((........))))))))...)).)))..((((((((((((((.........)))).....)))))))))).......)))))))))))))))))).((((((..(((....(((((((((.....(((((((......(((
.(((........))))))....((((.(((((((((....)).....(((((..(((((........((((((((((((.((((.....((((((.(((.....))).)))))).......))))..))))))).)))))........)))))..))..))))))))))...)))))))))))......)))))))))))
)))))))....)))))))))))))))(((((.........))))))))))))...)))))))......)))))))).))).))))).........(((((((............(((.............)))..(((...((((.....))))....)))(((....)))....))))))).......((((((.....
.............)))))).......))))).........)).))))))))))))))))......(((((.((..........))))))).........(((..........((((.((.((((........)))))).))))............)))....(((.....))))))).))))...)))))))))..)).)
))).......
You can download the minimum free energy (MFE) structure in
II. Results for thermodynamic ensemble prediction
1. The free energy of the thermodynamic ensemble is [-540.53] kcal/mol.
2. The frequency of mfe structure in ensemble 6.91323e-33.
3. The ensemble diversity 624.13.
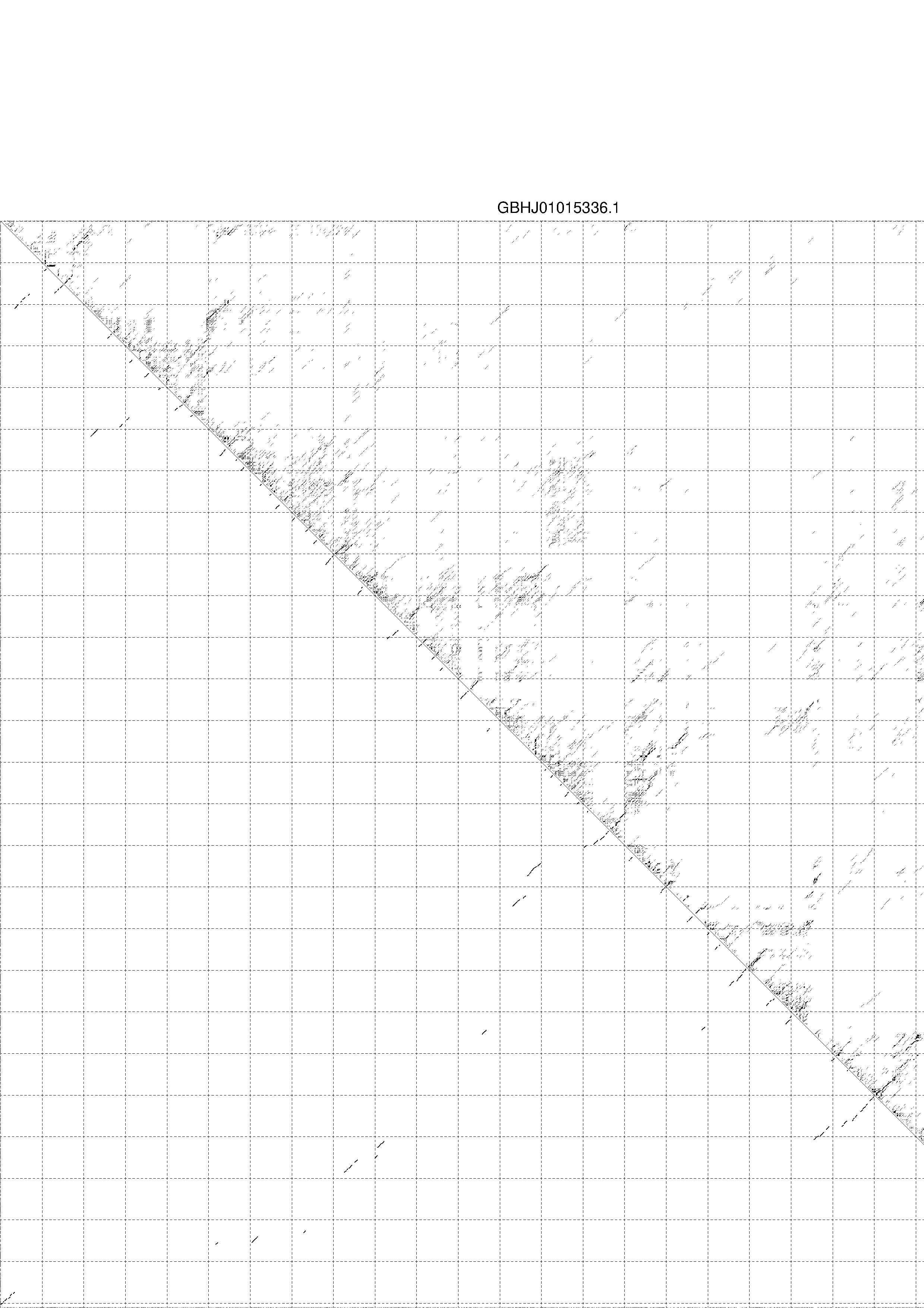
4. You may look at the dot plot containing the base pair probabilities [below]
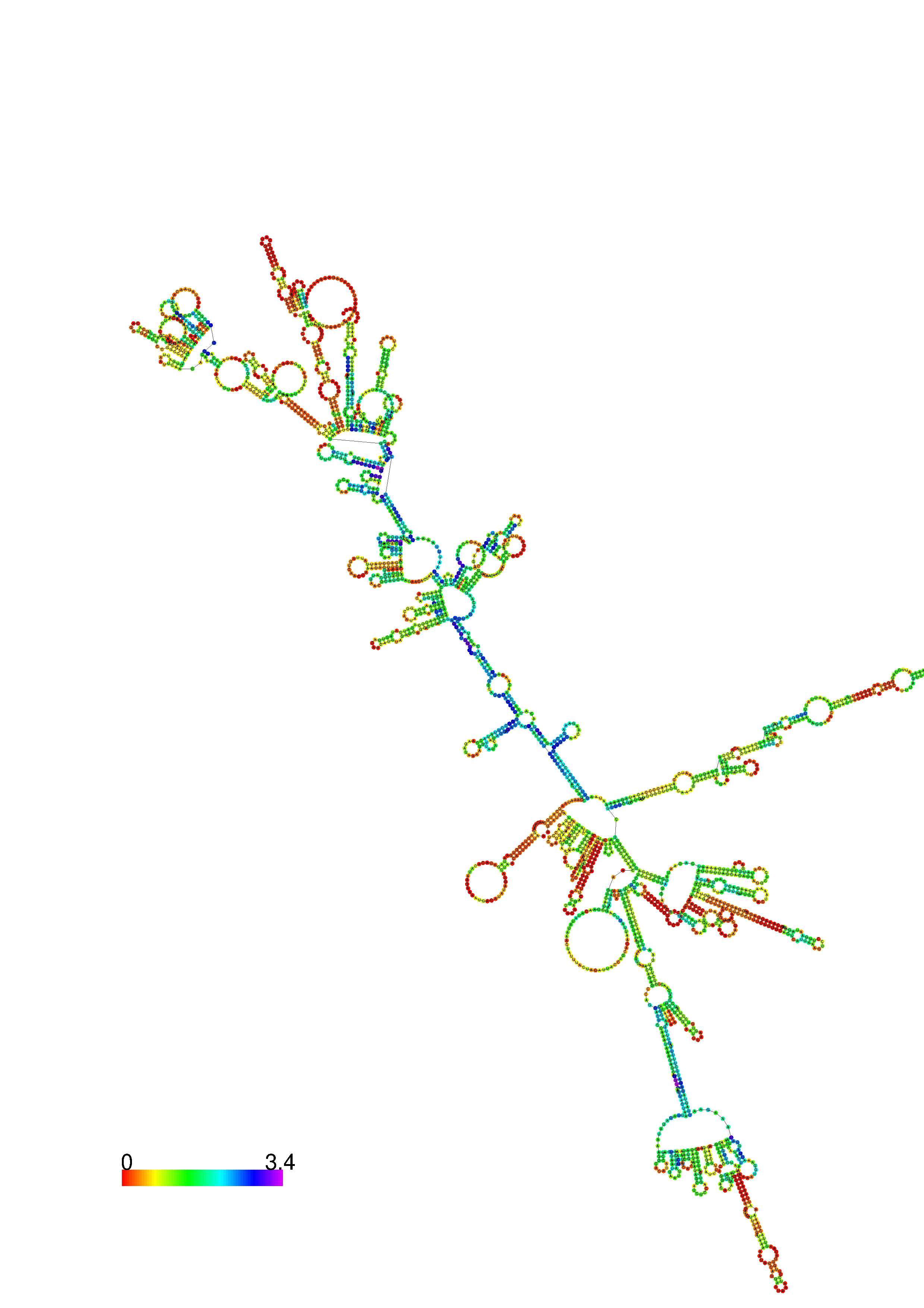
III. Graphical output
1. A note on base-pairing probabilities:--
The structure below is colored by base-pairing probabilities. For unpaired regions the color denotes the probability of being unpaired. The reliability information of RNA secondary structure
plot in the form of color annotation, Positional entropy ranging from red (low entropy, well-defined) via green to blue and
violet (high entropy, ill-defined).
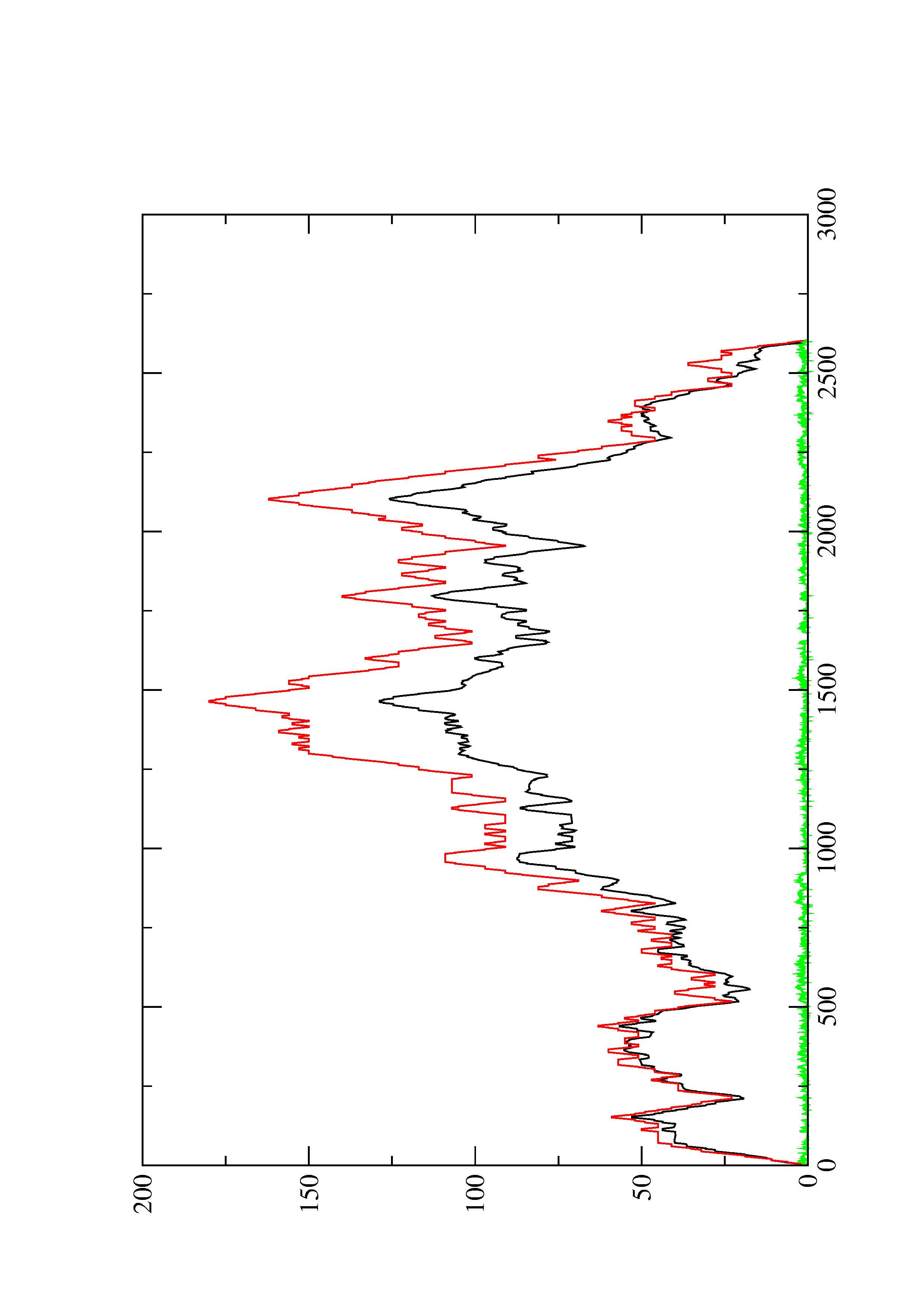
2. Here you find a mountain plot representation of the MFE structure, the thermodynamic ensemble of RNA structures, and the centroid structure. Additionally we present the positional entropy for each position. The resulting plot shows three curves, two mountain plots derived from the MFE structure (red) and the pairing probabilities (black) and a positional entropy curve (green). Well-defined regions are identified by low entropy.