lncRNA Structure Prediction
Species Name : Withania somnifera Sequence Id : GBHJ01015816.1
I. Result for minimum free energy prediction
The optimal secondary structure in dot-bracket notation with a minimum free energy of (-526.50) is given below.
1. Sequence-
CAGCAAACCAAAUAAACCCUAAAUUCUCUCUAGCCGCCGCCAUAUUUUCCUCUUCUACCC
UGCGAUUUCAAGCCCCAAGGCGAGAACCUAAAGGCAGUUAGAGGGUUAUUCUUGACAAAU
CUCGUCUUGUACGAACAAAAAGGGUAAUAUUCCUCUUUUCUUAUUGAUUUUCCUUCAAAA
UUUCGUACAAUCCAGCUCAUACACCUUACGUUCAAAAUUUGGAAGUUUUACUCCCAAAAU
CUCAAAUCAUUACUCCUAUAGAUACCCACUAUUCAUCAGUUAAGGGUAGCAGUUUUCUGG
CUGAGAGAUUUUGUUGAUUUCCUUGUGCGCAAUAGCUUGCUAAAACUGGAUUGGCUUGUU
UGUCCAUCAUUUUCAAAGUUAUUGCUAAUGAAGUAGCUUCGGAAGGUCAUGAUGUUUGUU
UCUACUGUCCUGAAGAAUUAAGAUUUGAGUCAAUAUCCUCGUGAUAACCCGCCAAGACUU
UUUAUGUGGUUUUUUUCACACCAAGAUAGACUUUUUGGCUUAAUAAAGUGAGGAUCUGUU
UUCUUGCGUUAAGAUCAAAGCUGAUGGCAUGUUCAAGUAUUCAGAUUACUUGAUAUUGGU
AUCUUGUAACCUGAUUUAGCCUGUAGCAGUUGAUAUAACGUGCUUAGGGGUAAGAGCUUU
UCUGGCUCGAAUUCAGCAGAAUUUGUCAGUUUUUAUAGCUUGAUUAGUAUUGCAGGUUAU
GUUGAAGGCUCUUAGUCGUCAUUCCCUAGAAUAUGCUUGGUCUUACCUGAUUUAAAGAAU
CUGCAGUGAAUGUUUAGGAUCAACUUGCCUUAGCUGUUGAUUUUUCCUUGAUCAAGUUUG
AGUCUUUCAUGAGAAGGAGAAACUGAAAUUCAAGAGGCGAGGCUGCCCAGUUUGAACUUU
UAAAGUUCAAGCUGCCCCUCUAUUUUGUAAUCAGUGCUGUAUACCAAGAGUUCCCGUGAU
AAGGGAAUAAGUUAAAAAAUUGAUUCUUUGUGAUCUUUCAUGGUGUUAUUUACUGUUGUU
UGUCGUUUUACAACCUAGAUAUGUGUACUUAGGUUAAAGAUGGUUAAUUCAAUGGUUUCU
GUUAUAUUUUGACUGUAGUUGGUGGCUUAGAAAUAUAAAUAGGUGCAUAAAGAGGAAGAA
AUCUCUGAAUGAUAAGUGUGGCGCAGCAUAUUUCAUGCUUUACCUCACGCUAAAAUGGAG
CCUUUACGACUAAAUUGAGUACAUGCUGGUGUUGCUUGUAUAGCCUGUUUAAAAGAAACU
UGUAUGCACUAGUUGGUUUAAAGUCAUGUUGAACAAAACAUGUGUUAAAAAUUGAGUUGC
CUUGAACAAAGGUUGUUGCUUACCCACCCUUUCCAUAUGAACCUUUUCCACUGUUCAUAU
GGCAUUUCUAGGUGUUGCUGCCCCCUAUCCUAGUCUUGUUUGGCACUUUGAUUAGGCUUG
ACUUUAGGGGAUUUAUAUUUGUGAUAUCUUUUGUUAAGGUAGAUUGACUCUUUGCGAGAA
GCGAUUGUUUAAAAUGUGUCCAACCUUUGUGCAUUGGCUUGAGCUUUUAUGUUUGAUUGA
UACCGGAAGUAAAGAAAGACUCCGUUUUGUUAUGGUUUAAAGAGCUUUCACGUGCAUGUU
UGCCUGCUGUUUUCCUUUUUUUGAUAAGUAUGUAAUUUGGAAUGAUAAGCCAAUGGACUG
UGACUUUGCUUUAACUUGCCUUUAAAAUUCUCACACCUUUUGGUUUUACAUUCAAAUAUU
CGUUUAUUAUUUGUUUAAACUCUUGUGCUAUAUGUUUUAAUUGACACUUGCUUUGCAUUA
AGUUUGCUUAUGUCUCUAAAUUACGUCAAUUGGCGAUUAAUUUUUCUCUUCUUUCUUUGC
AUACUCCUCCAUCCGAGUCCAAUUUGGACAUCAUCCUCCUUCGUGUCCUAUCUCUUUGAG
UCAAACCUCCUUCACCCAAGUUCUCUGAAAAUAAGAAAUGGCGUAAGAGCCAUACGUGAG
AAGCGAAGAAAAGGAAGCUAAUAUACAGGCUAGGCCAUGAAGACGGAGCAUAAGUUAAAA
GGAGCUAAACCCAAAAAAACAACUUUCGUAGUCCUAAGUUGGACACUAGUAGGGGUUUGG
GCCUAAGUGCCUCAACCUAAUUUCUUCUCCUCUUUAUUUAAUUUCUUGUAAUAUUUGUUG
UGCAAAAAUCCUUUUGUUUAUACUUGGACGAAUCCUC
2. MFE structure-
..(((...((((((...((((.......(((((((((((.........(((((.....(((.....(((..(((....)))..))).....))).....)))))((((....))))..........(((((((((..(((((((........)))))))....((((.......))))...)))))))))..((((....
.................(((((.(((...))).)))))................((((..((((..(((((((((((((.(((((.(((((...((((((((((.((((((((((((.(((((((.((((((((((((...((((...(((.(((......))).))).))))..))))))))))....((((...))))
((...(((((((....((((.((...(((.(((....))).)))...))..))))...)))))))...))))))))........(((((......)))))((((((......))))))((((((...(((((((......)))))))))))))((((..((.((((.((...(((((((.......)))))))...)).)
)))..))....))))(((.(((..((((((((...)))).))))..))).)))(((((.....)))))))).)))))))))))).))))....(((((((((..........))))))))).(((.((((...)))).)))....))))))..))).)).))))).))))).....))))....))))..))))))))((
((((..((....)).))))))((((((((..(((....(((.....)))))).))))))))(((((....(((((...((((.....((((((((((((...))))))))))))..))))..)))))...))))))))).((((((.((((((((.......)))))).............((((......))))..)).
)))))).....(((((((....(((((((..(((((((..........))))))).)))))))......)))))))....((((.....))))......))))))).))))........))))...((((((((((((((((((((.........(((.((...(((((.....)))))...)).)))........))))
........(((......)))...(((((((((((((((((.(((..((((((..(((((.((((.((((.((..((((((((..((((((......))))))................(((.((.((((((((((..((.((((........((((((((((..........))))))))))........))))..))..
.((((((....((((..((((((((...)).))))))..)))).))))))...(((((((..((((((((((((.(((((......)).))).))))))...)).))))..)))))))..)))))))))).))..))).(((((....((((((((.((..(((((((((((((.(((.....((...(((((((((((.
...(((((((....((...((((.((((..((..........))..)))).))))..))...))).))))...)))))))))))...)).....)))..)))))............))))))))....)).))))))))..))))).......)))))))))).)))))))).)))))..)))).))..))).))))...
)))..(((((((.(((.......((((...((((((........))((((.(((((((((..((((........)))).......((((((...........)))))).........(((......))).(((((........(((.......))).(((((......)))))..)))))..)))))))))))))((((.
.......))))..))))....)))))))))))))).....((........)).......((((((..........)))))))))))))))).(((((.((((......)))).))))).))))))..))))))))))..............))))))...))).........((((((((....)))))))).....
You can download the minimum free energy (MFE) structure in
II. Results for thermodynamic ensemble prediction
1. The free energy of the thermodynamic ensemble is [-567.76] kcal/mol.
2. The frequency of mfe structure in ensemble 8.42825e-30.
3. The ensemble diversity 637.77.
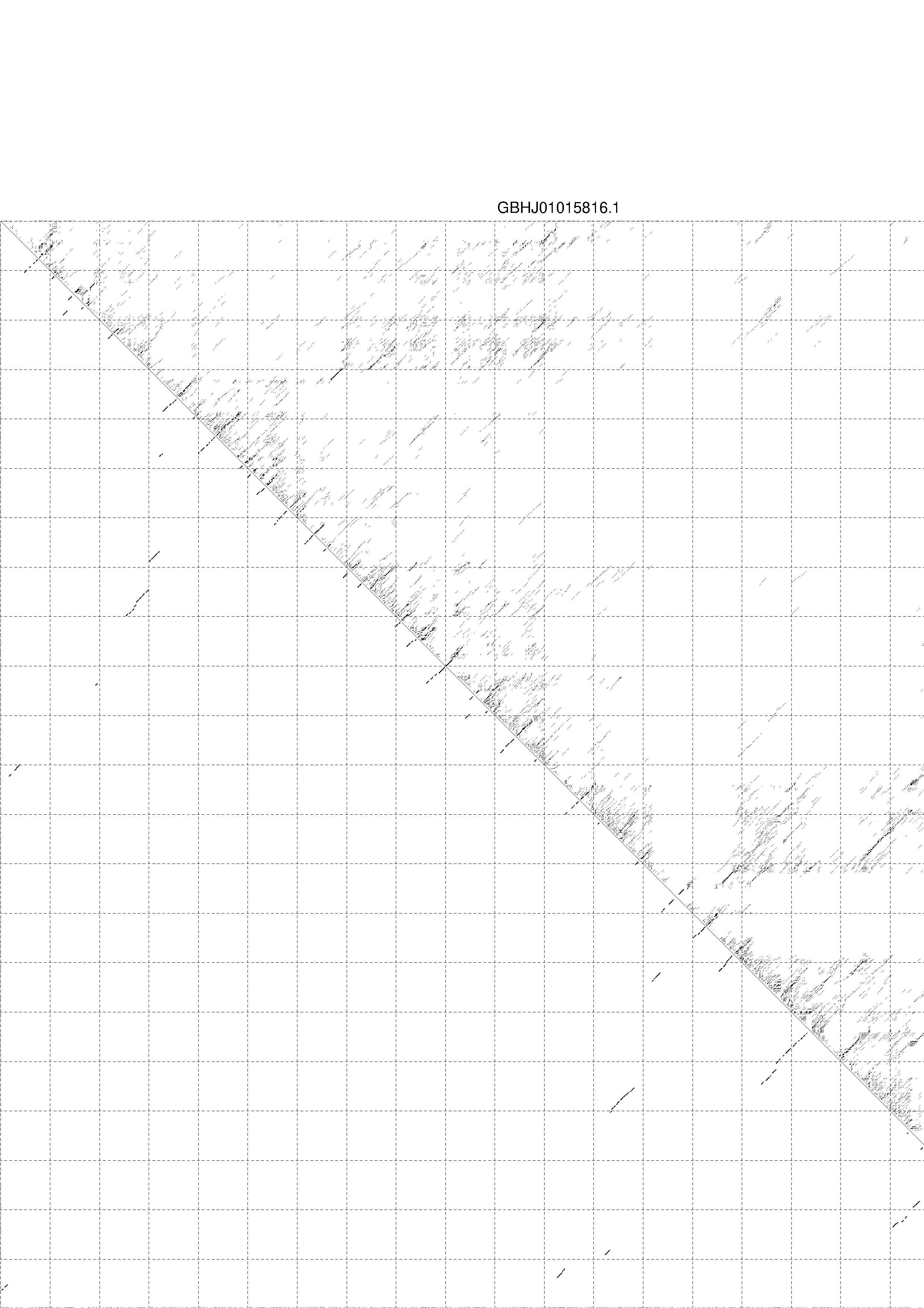
4. You may look at the dot plot containing the base pair probabilities [below]
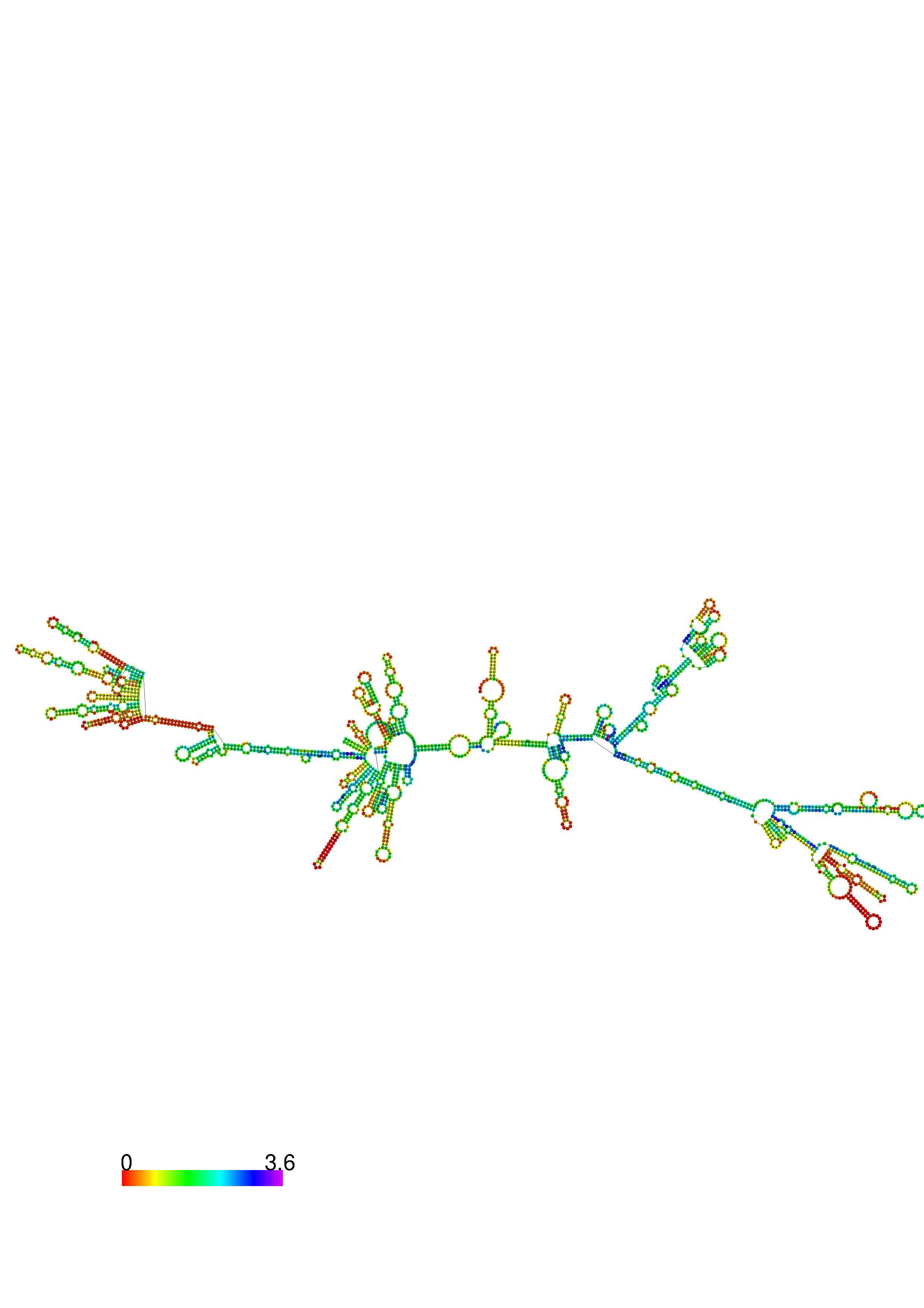
III. Graphical output
1. A note on base-pairing probabilities:--
The structure below is colored by base-pairing probabilities. For unpaired regions the color denotes the probability of being unpaired. The reliability information of RNA secondary structure
plot in the form of color annotation, Positional entropy ranging from red (low entropy, well-defined) via green to blue and
violet (high entropy, ill-defined).
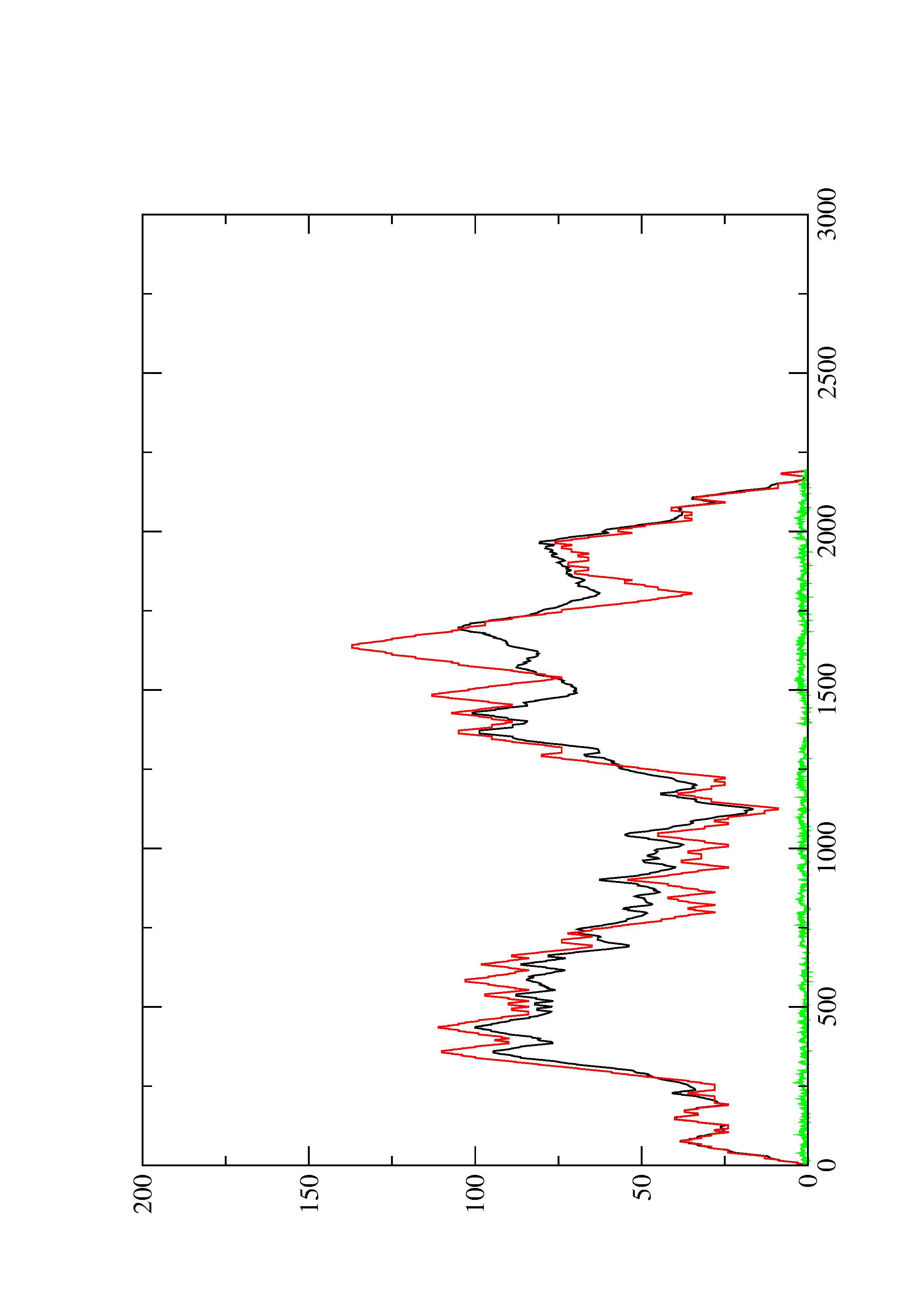
2. Here you find a mountain plot representation of the MFE structure, the thermodynamic ensemble of RNA structures, and the centroid structure. Additionally we present the positional entropy for each position. The resulting plot shows three curves, two mountain plots derived from the MFE structure (red) and the pairing probabilities (black) and a positional entropy curve (green). Well-defined regions are identified by low entropy.