lncRNA Structure Prediction
Species Name : Withania somnifera Sequence Id : GBHJ01015920.1
I. Result for minimum free energy prediction
The optimal secondary structure in dot-bracket notation with a minimum free energy of (-670.40) is given below.
1. Sequence-
UUUUUGGACUGUUUUCCUCAAUCUAAAAAAUGCUUACUGUUUUCUCUCUGUUUGUUGUGC
UGAAUUUAUCACCUCAUGCUCAAACCCCUUCCUUAAACCAUCAUGUCGUCUACCCAUUUC
UUUUCUUCUACUCUAUAUAAAAUUUUUUUUGGCUCUCUUCUCUUGAUGACAUAAGAGACA
GUCUUAAGCUAGAUAAUGUUGGUAUAGGACUUGUCUUCUUUAAAUCGUAUCUCCAAUAAU
AUAAGAAUGAGAUACUUGCCUUAAUAAUUCUUGAAGAUAUUCUAGAUUACCCUUGAAAUG
ACUAUGUGUUAUGUAAAAGACCAACCUAAAUGCCCUGCCUAAUUUAUGUGGAAUUAGUCU
UAUAUCAAGCUACCUAUAUCAAUGUUCUUUCUCUCUUCUCUCUUUUCAAACAUGUCCAUA
AUUGAAAGUAAUUAUCUUACCUACAUCAUUUGGUUCUUUGAUGAUACGGCAGUAAUGGAA
GCAUGCUAAUGUUAAUUUGUCUUUUAGCUGAUAAAGUUUGUAAUUGACUGUAUAGAGAGU
AUUGACAACAUGAUUUCCAGUCUACUUCUUUACCAACAUGUUGACUUACUUAGAGUGAUU
AUGCACACCUAUUAAGGGAAAUUAUACUUAAGUUUCUAUGAGAAGUUGAGCAUGUUAUGU
ACUCUUUUCCAAGUGUUUUGCCCCUGUUUCUUGAACCUGACUAUGUGCUGGUCUUGUACA
AUGCUUCCUGGCUCUAUUUGCUAAUCUAUUUGACCUUAAAUUCAUGUAUGGUUAGAUGUA
GUAGCAUCUUGGAGUAUGUCUUUUCAUAUUAAUGCUAGUUUACUUACUCAAGUGUUGAAU
GAAGCUUGUUGUUAGCUGAUUUGGUUAUGUAUGACAUCCUUAAACCCUCCGUCCCAUAAC
AGUAUAACAAUACAUGCCGAAGUAUGACUAAACCAUGCAUCUAUAUAAGCUGUAUCAUUA
GUUAAAUAACCAUGAUCCCUCUUCGAUAUUUCUUAGUUUAUCCACCUGAUCUUACCCUCU
AAUGCAUGCACAUGAUCCCAUUGAGCUAUAUUGAGUAAAUUUUUACAACUUUUGUAGAUU
AUGUAUCUUUAUUUGGAAACUAGAAUGGUAUACUUGUUAAAUUAUCCCUGAAGCUAUCUA
GGAAAUGCUAGUAAUUGCAUCACCCUUGAUAUGAUGUUAAAAGUAUAGUCAUCAUAUAGA
AUAAAACAUGGCUCUGUUAACUUUGUGGCUUUAAAUGAUUUUCCAUAUAGUUUCUUGCAG
GUGGUGCCUUGCUAUUAUGAGCUCUUGACUGGCCUUUUUGCAGUAUUCUUCAAAGCUUGC
UAAUUUGCUUGUUGCUUGUUAUGAGCUGUAUUACGUAAUGUAUCAUGCAUGCUUUAAUGU
GUAUUGAUUUGUAUGUAACUUGUCUAUGUGGUUACCUCUAACAACUAGCCUGUGUCAUGC
UCACUUUGGCUUAACUAACUAACAUUGAUAACAUUUCUGCACGACCACUGUUAUUGCUUA
AUUUUUUUAGGUUGCCUAAAUUGAAGCUAAAAGGAGAUAACAUUAGGUCUAAAUGCCAUG
ACUAUAAUCUGCCUUAUUUCCAGUUAUCUCUAUGUUUCCUAGUUGUUUUCCUAAAUGUUG
UCUGACAUAGACUUCAUGCUAACUGAUUGCUUGCAUAAUGAAAGAUGCAUCUGGUCAUUC
AUAAUCUCCUUAUUGCCUGCUAACCUAUUGAGAGUCAGUAAUAGUUUUAUAUUUACCCAU
CUAUGUGUGUAGUUGAUCAAGUCUUAGUGAUGUUUCACAUGAAUUAUAAUAGAUUGUUGA
AGCUGAUGUAUCAUGUGAAUAUAGUUUAUUUUGGUACCUUGAAUGUUGGUAUAACUUACU
CACUGUUUCCUCUCUUAAAGUCAACAUGCAUCCUGAUUGCUUUUCUCUUAUUAAAAUCUC
AUAUAUUGAACUUUCUUGCCCUCUUUUCAAUCACACUUGUAUGCAAGUUCUGAAUGAAUG
AUUGAUUUAGAGUAUAUUUUCCUUAGACCUUAAUUUUUUUAGACAUGUAAUGCUAUACUG
GCUUUUACCUGUUAUUCCCUGCUAUGUCACUAGCCUCAUCUCUAAAUGUUUGCUUGCUGU
CUUGGAGGUGUCGACUCAAAGAUUUGGCCUGGCUUGUGUAUAUUUUCUCUUGUUGCGUAU
AGUCUUAUCAUUAUGCGCCCAAAAUUCAAAUCCCUUACCUCCACCAUACAUGUACUACUG
GCCUCGUAUAUACUAAUUUUAUUUUGUGUGAUCCCAUGCAAGUCUGCUCCUUUAGUGUCA
ACAUAAUAUACUAGGCUAAGAGUUAAAAGGAAUGUAAAAAUGUGGAGUCGAAGUGCUCAU
CUUCAUUCACUGUAUUUGCUUAAGGUUAAGGUCUAAAUGCAUGUGUUUACUUCGUACAUU
UGAGAUUGAUGGGUCUGUUACUUUGCAUAAGGUAGGCCUAGUCUAAUAAGUGUUAAGUGC
UAGUAGUUGUAAUUUAGAUCUGUAUUUGCUUCAAUUAUGUUUGCUAUUGCUUAUCUACAU
AUGAUUUUUUGUGUCAUAUGUAUAGUAAAGACUAAAUUGAAGUCGUAGUUUUAUUAUCAC
CCUUGCAUACUAAAGUAGUGUUAAUAAAGUAUAGACUAAUUGGGCUGCAUGUGUAUUUUC
UUUUUUGAAUAUGUGUUUUUGCCAGGUCCUAAUCUGAAGCGGAUUGGUCCAUGAUAUGGG
CAUGCUUUCAGACUACCUUAGCCUAUUUUUUAUUUGGGUCCUUAGUUUCUGGUUUUUCCC
AACUAGUUCUUCCAUUGACUCGGCCCAUAAUUGUUUUUUUUAUGAAUCAAAUGUUUCUCU
AUAAUUAUUUAGCCUUAUUUAUAUUCAUUUGUGUUACUUAUUUCUAUCGUUCCUAAUCCU
UUUUCCCUCUUUGCGUGAGUGAUCUUGGGGUUCAGAAUCCUCUUGCAUUGCCUUGCGGGC
CACGGCUCCAGUUCAAGAGUUGAUUUUGUUCGACAGGAAACCGAGAAGAGUUUAUCCGUU
CUUCCUUCUUCCGUUUUGUCGUGUUUUAUCUUAUGUAUUUAUUAUUAUGAUGGUACGGAA
UCAAGGGAUAUCCCAGCAUACGAUGAGUCAUCUCUGGGAUUUUUAGUAUUAACUGGAUGU
UCC
2. MFE structure-
((((((((.((.......)).))))))))................(((...((((.((((((....((((...(((.(((..(((....((.........((((((((((.((......(((.((..((((...((((((((((.((.(((((((((.(((((((......)))))))..((((......))))((((((
((((((((.((((.............(((((((...............))))))).............(((.(((....))).))).....(((...(((((.....)))))....)))...................(((((((.....)))))))........))))...))))))))))))))..............
...((((((............))))))(((.(((((..(((..........))).....))))))))((((((((.((((((((....)))))..((((((........))))))....((((...(((....((((((((((((((..(((((((((((((.((..............)).))))..(((((.(((...
.....)))...)))))))))))))).(((((.(((((...))))))))))..))))).)))))))))....)))...))))..........((((..((((((.((((..(((((.....((((......((((((..(((((...(((((((((.((........)).)))))))))...)))))...)))))).....
..(((((.(((((((((((......)))..)))))))))))))(((.((((((((........((((((...(((...............))).)))))).(((((((((((((((..((((((((.......((((.(((((((.(((.((((((((..(((....))).))))......((((((((..((((((...
.......((((......................))))..))))))..))))))))........)))).))).)))))))))))...........((((((((..((((..........(((((((...(((((((((...((..((((........))))..))....(((((((((((......))).))))))))...
....((((......))))......))))))))).))))))).)))).)))))))).....(((((((.((((((.(((.(((((.((((.(((....(((.((....)).)))(((..((......))..)))))).)))).)))))))).)).)))).))))))).))))))))..)))))))))..))))))((((((
.........))))))..)))))))).)))((((((((.((..(((((((..((((((...(((((.((((((......(((.(((....)))..))).......((((((.((..........)).))))))((((((((.((((((((..........)))).)))).....)))))))).))))))...)))))...)
))))).....))))).)).)).)))))))).......((.........))..)))).....))))))))).)))))))))).....)))))))))))............))))))))).)).))))))))))................))))..))..))).....))))))....)))))).........))..)))..
)))))).((((((.((((((...))))))..))))))...(((((..((((.(((((.(((...(((((((.((.((((((((((..(((.......))).........................(((((((.....(((......((((((.((..((((((.(((((((((((.((...)).))))))).........
..(((((((((((((((....((((.((((.((((...))))..)))).)))).........((((.(((.((((..((((((((..((......))....))))))))(((((.((...)).)))))..)))).))))))).......(((..(((((((((......))))))))).)))................((
((((..(((((..(((..((((((.(((((((((((......((((((((....)))))))).........))))).........)))))).))))))..)))........))))).....))))))..((((.......)))).................)))))))))))))))............))))))))))..
)).))))))..(((((((.....((((..(((..((((.((((.....((((..(((.((((((.........((((.((((((((((((((((((..((((....(((((....((((((((((.......)))))))))).)))))))))..)))))))))........(((((.(((.(((........)))..)))
.))))))))))))))))))...(((.((((((((....((......))))))))))....))).((.((.((((((...)))))))).))......((((...(((...((((((.....(((((.........)))))...))))))..)))....)))))))))).))).)))))))).))))(((((.........)
))))...((((((......(((((.((........)).)))))..)))))).................((((..((((((...((.(((.......))).))....))))))..)))))))..)))).......)))))))..)))...)))))))..))))))))))...)).))))))).)))..)))))))))))))
)..((((.((((((.((((((((......................)))))))).))))))..))))))))...)))))))))).)))........(((((.((((((....)))))).)))))
You can download the minimum free energy (MFE) structure in
II. Results for thermodynamic ensemble prediction
1. The free energy of the thermodynamic ensemble is [-723.58] kcal/mol.
2. The frequency of mfe structure in ensemble 3.33434e-38.
3. The ensemble diversity 907.17.
4. You may look at the dot plot containing the base pair probabilities [below]
III. Graphical output
1. A note on base-pairing probabilities:--
The structure below is colored by base-pairing probabilities. For unpaired regions the color denotes the probability of being unpaired. The reliability information of RNA secondary structure
plot in the form of color annotation, Positional entropy ranging from red (low entropy, well-defined) via green to blue and
violet (high entropy, ill-defined).
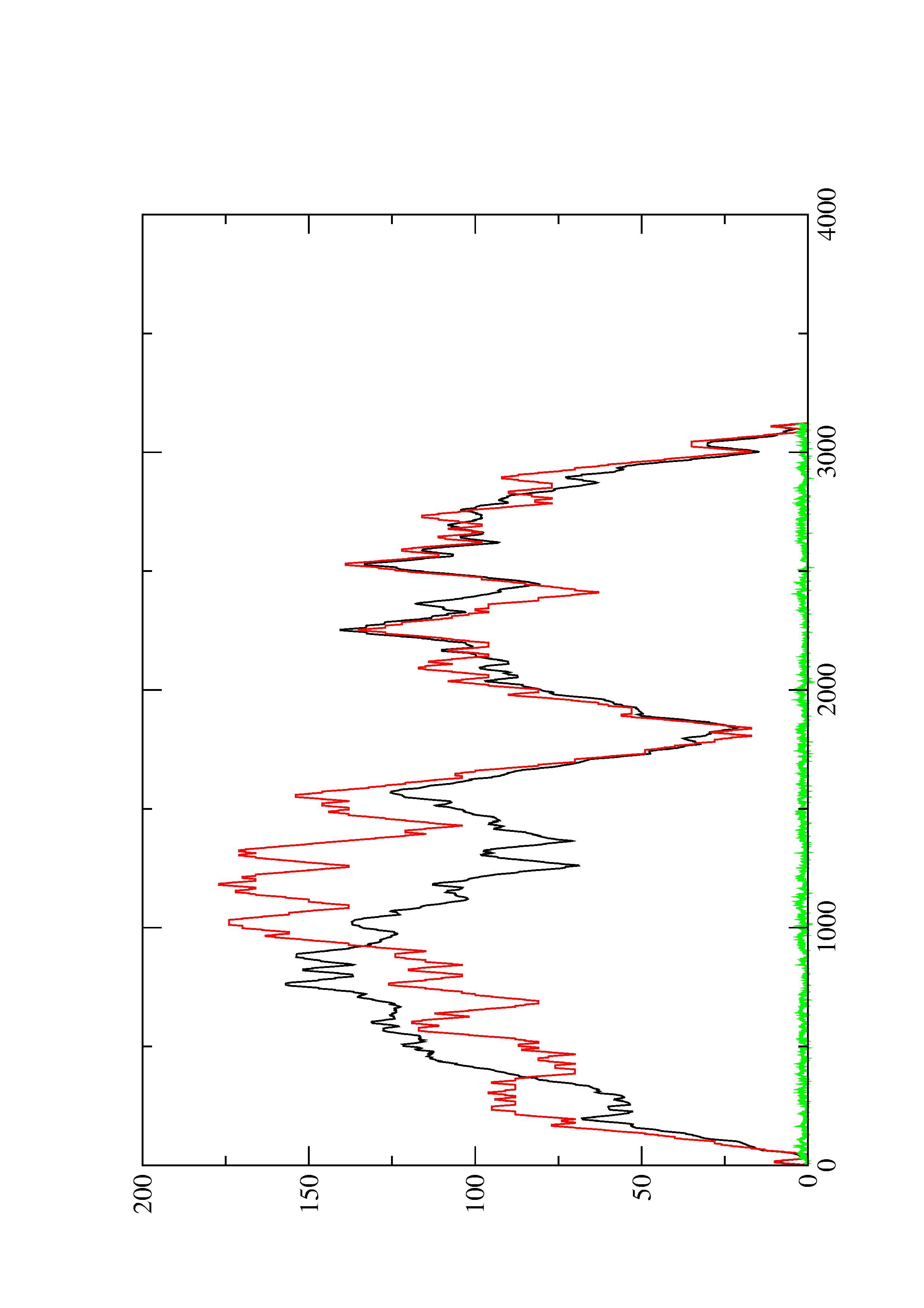
2. Here you find a mountain plot representation of the MFE structure, the thermodynamic ensemble of RNA structures, and the centroid structure. Additionally we present the positional entropy for each position. The resulting plot shows three curves, two mountain plots derived from the MFE structure (red) and the pairing probabilities (black) and a positional entropy curve (green). Well-defined regions are identified by low entropy.