lncRNA Structure Prediction
Species Name : Withania somnifera Sequence Id : GBHJ01016624.1
I. Result for minimum free energy prediction
The optimal secondary structure in dot-bracket notation with a minimum free energy of (-478.11) is given below.
1. Sequence-
AAAUUACAUAGUCAAAUAUCUUUCACUCACAAUUGGGAUAGGAGAAAUGCAUUAGAAGAG
CAAAAAACAACAAGUUAUUCAUAUACUACUUAGAUUUUGGUAUGAUGUAAAAAUUAUGAG
UACUAUAAUUCCAUUAAAACUUCAAAUUUAAUUUAAUAUUGAGUGAGUGAUUACUGCUGG
CUUAACAAAAUACAUAGACUUAAGGGAAUUGCUUCAGUGGUUACUAAAAGAAGAUUAUUA
CCAAACUUAACCCUUUUUUUACAAAACUAUAAAUGCAUUCACCCUUUAAGUGUUACAUCU
UACAUUAAAAUAAACAUGAUUCUUUAUAGAUCUAAUCCUAAAUCAACCUAGGCAUAUGAU
ACCAAUUCAAAUCAUAUAGCACACAAAAUUGUCUUGAGGCCCAUUUCACUUUCUCUUAAA
ACCACAUUGUCAAGAACGAAGGAUCAUAAUAUAGAAGACAAUUAUGAAAAGCUUAGCCAA
UUGGAAAUUGAAAGACACAGUUAAUAUAUUAAUACUAGUAACAUUCAACAUAAUGAAACC
AAUAGGAUAAGAAACCAACUACCAGGAACAAAUGUAAAGUCACACAACAAGCUGGAAAAA
AGUAUAACACUAGACUUACACUAUCAGUACAAAAAGGAGAAUAUUAAGUGACAAAAGCUC
UUAAAAUAGUCUCUAGGACAUUUGAAUGUCCUAUGUCCCUCUGCCCUUUAAAUAUCAAAC
UUUUGGAUUCAUAGGAAAUAGAUUGAACCCCAUAUGAAUCCACACUCCUCUAUUUAGGUA
UUCUUAUAAGAUGUGGGACUUCUUGGAUAAAUUAGUAGAAGAUUGCCAUCGGGGCUUAUC
CAACUCACAUCUUACAAGUCUUCCACUUAUUUUGCACUCAUGUAGGUCCUUAUGCAGUUA
GGUGAGGGAGGAAUGAGGUAAAAAAGUCAUAUGAACCUACUACAUGUAUUCAAAAUAGUU
UUGAAACCAAAAAUAGACUCUUUACAGAUCACAUAGAAAGUAAAAUGACCAUACAUGGGC
UGGCUGCUAAAACAAGAACUGGGGUUGGGCUAAGAGGCUUCAAUUUAUGAUUUGAGGAUG
UCAAGAAGAUGACUUCAAGAUUAGAUUGAAACAUGGGAUGAGCGAAAUUAGAAUUUACUC
CCUUACCCUUCAAAUCAAAAACCACCUUAAUGUCUCAUUUUCUAUUAAAAACAAUAUUUU
UCAAAAGAGUUUGGAAAAAAGAAAUAUGGAACAAAAACAGUUUUUCACUUAGAUGUAAGC
AAAGAUAAUAACACACCUAACUGGAAUCCGUAUAGCAGAUUAAUCCUAAUGAAACUCUUA
CCAAACUCACGUUUAAAGUAAGAACUCCUAUAAUUUGUAAAAUUAACAGGAUAGCAAUUU
UCAACUACAACUAACAUAGCAUUUUACAAAUAUGACAUCAAACAAGUGGAACAAAAGACU
UAACAACAAGUAUAAACAAAUAAGAGUACAUAAAUACUAGAACCACCUCCUAUAACCCCC
ACAGAUCAAGCUUAAAUCCAAGUCAAGCCAAGCAAUUCUCAUAACCAAGUCAAACUUCCC
AUUAUUAGAACUAAAACAAAAGAACGACUUGAUGAACUAAGAUUACACAACCUUCGUAUA
CAAAUCGACAGUUUUAAGAUAACUUUAAGAUAAUUAGCAAGACUAAAAGUGUACCUGUUC
AGGAGCAUGAAUCAAAGUCAAAGAAUUAAAAAAAAACCUUCGACUAGGGAAAAAAGGCCA
CUAAGCUCUCCAGCCUUCUAGACCUUCUUAAAUGAGUACAAGGAAAGACCCUUUAAGAGA
AAUACUCAAAGGAAAUACUCAAAGGGGUCGAGGAUCUCUCCGAUUUUACCUCAACUAAAC
AUCAUUUAGAUAAACACUUCAGGGGUUUAACCCUAAUAGAACUAACAAAUAAUGAACCUC
AAGAACACCACACUAAUUAUUCAGUAAACUAUGUGAAAUCCUCGAGAUCGUAGGUCUCGU
AUUACUGCAUAUGAUUAAGGAAGAAGCCUAUUAUUUCAAAACCCUAUAGGUAUUGCUUAA
UACUGAAACUAUCAUCACUACUAUUCACUUCCUUAAGAUUAUCAAAGAACUGCUCGAAAG
GUAAAGGGGAAAAGGUUUAAAAAGAGAAGGGAAACUUAGGCAACAAGCUCGAUUUUAGGG
GGUAAAACAAACUCUGAAAAUAAAAAAACCCAAAUCAAAACAUCCAUCAAUCGAAACCAA
UUAAAAAAACAUUAAAAUCAAAAGGGUUUUAAAAUUCAAAUUUUAGCCAGGGGAUGGGAA
UGAUAGGCCAAUCAUUGGCCUUAAAGGUGUACAAAACAUAUAAGGAAGACAUCAUCACAA
AAUACCUUAUCAGAGGGGAUUCUAUUCUUCAUAGGGUUCAAGGACACUCCUACAAUGACU
CCAAAUUCUAUGGACAUUUACAUGCCAUGAUGGAAAAAGAGUACAAGGAUUGAAAGGCUC
UCGAACAUUCAAGAAAUAUCAAGGGAAAGAUGGAAGGGAUCAUUGUGGCCGUAUGAGAGG
GAAAU
2. MFE structure-
...((((.((((..(((((((((((.((.(((((((..(((((...........((((((((.........((((............))))..(((((((((.(((((((.(((((((.....)))))))..((((((........))))))......((((((.((((.....)))).))))))............(((
((((((...(((....((((((..(((((((((...................)))))))))...))))))....))).....))))))))).))))))).)))..)))))).....)).))))))..........))))).......(((((((((((((..........)))))))......((.(((((((((.....
........(((((((((...((......)).))))..)))))............))))))))).))....)))))))))))))))...)))))))...........))))...)))))))).......((((((((..((.((((..((((..........((((.......(((......))).......)))).....
(((.....)))...))))..))))...........(((((...(((((...........))))).....)))))(((((((....)))))))...(((.(((.(((((.............((((((.((((((((......((((((((..((((((......(((((((....(((((((....(((((((((((...
..((((((((..............(((.....))))))))))))))))))))))...(((((((((((.((((((.(((((.....(((((((........)))))))....)))))))))))))))...............(((((.(((((..((((((((((..((......(((((((..(((((((..(((..((
(...(((......)))..)))..)))...............((((((((((....))).))))))).))))))))))).))).......))..))))))))))..))))))))))((.((((.((.............)).)))).))..............((((((....(((((((((((.((((((....((((((
(....((((((((..........(((((((............(((((.((((.(((................))).)))).))))))))))))......................((((((.((((....))))...)))))).........(((((((((((........(((................))).......
)))))))))))..............((((........((((......)))).............((((......))))....))))........((((((..(((((.(((((....((((((....((......(((((.....((((((...(((......................)))...)))))).........
........(((((((...(((...(((((((((((..(((((.((((((....((((.....)))).(((((...((((((((..(((.......((((.(((..............))).))))(((.....(((((..............))))).....)))..((((((((((.........((((((((..(((.
....((....)).....))).))))))))(((((((.....)))....))))......)).))))))))...........((((......)))).................)))..)))...)))))..))))).....((((((....((((((.......(((((((...)))))))........))))))((((((.
((...((((((..............)))))).)).))))))))))))........................)))))).)))))..))))))).))))...))).....))..)))))......)))))))...)))).))....))))).)))))..))))))....)))))))))))))))..(((((((.........
...........................................)))))))..........))))))..))))))))))).....(((((((...)))))))..))))))..............)))))))..........)))))))....))))))).........))))))))))))))......)))))...))).)
)))))...(((((.(((....))))))))(((.....(((((..............)))))....)))............)))))..))).))).))..)))))))).((........)).....
You can download the minimum free energy (MFE) structure in
II. Results for thermodynamic ensemble prediction
1. The free energy of the thermodynamic ensemble is [-523.86] kcal/mol.
2. The frequency of mfe structure in ensemble 5.78019e-33.
3. The ensemble diversity 543.70.
4. You may look at the dot plot containing the base pair probabilities [below]

III. Graphical output
1. A note on base-pairing probabilities:--
The structure below is colored by base-pairing probabilities. For unpaired regions the color denotes the probability of being unpaired. The reliability information of RNA secondary structure
plot in the form of color annotation, Positional entropy ranging from red (low entropy, well-defined) via green to blue and
violet (high entropy, ill-defined).
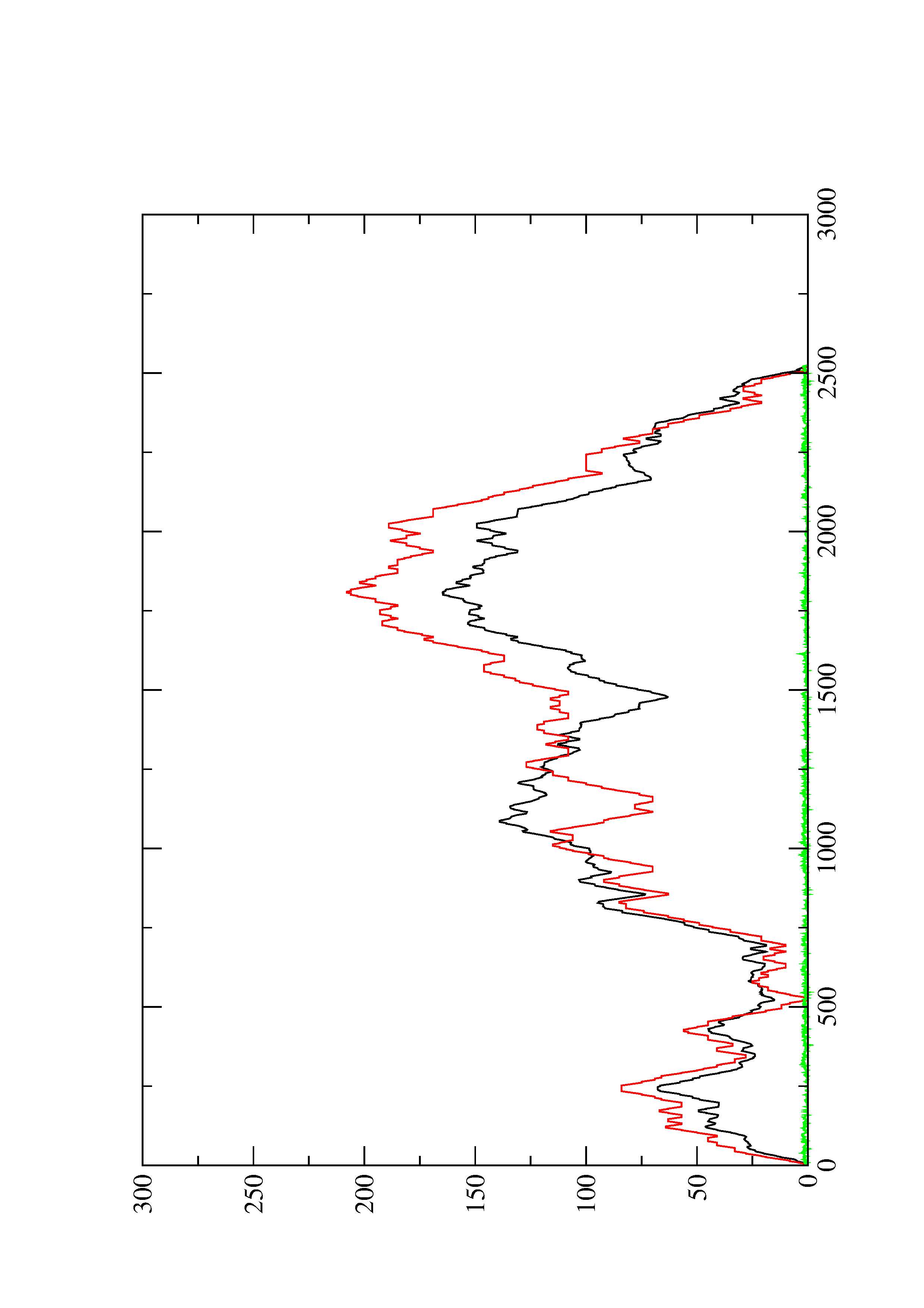
2. Here you find a mountain plot representation of the MFE structure, the thermodynamic ensemble of RNA structures, and the centroid structure. Additionally we present the positional entropy for each position. The resulting plot shows three curves, two mountain plots derived from the MFE structure (red) and the pairing probabilities (black) and a positional entropy curve (green). Well-defined regions are identified by low entropy.