lncRNA Structure Prediction
Species Name : Withania somnifera Sequence Id : GBHJ01016928.1
I. Result for minimum free energy prediction
The optimal secondary structure in dot-bracket notation with a minimum free energy of (-594.50) is given below.
1. Sequence-
GCCAUAUCUCCUUUCAAGACUCUCAUCUUUCUCUCGCCUCUCACUAUAACUCCGUCCAGC
AACCACAUCGCCACCGAUCACUGCUCCAGAAGAACCCGCUGACCAACACCCUCCACUCUU
CUUCUUCCUCUCUCUCAACAGCCACAAACACCCAAGCCGCUGUCGUGUUACCAUUUCCCA
CUGGAUAGCAGUCCUCAUCCAGGCCAGCACCCGACGCCGUCACUUCCACCACUGCAGAUC
UAGUUGGUGAAAUUAGAUCCAUCACCGCCGCUUUCUCGUCUUCCCUGAACACAAUUCUAU
CCGGACAGCACCACCGCUCCCACGAAAAUGACUCCGCCAUGCACUAUCAGCUCCACUACA
UUUUCUCUAAUUCUGGACGAACUCUGACUAAAACUCACCGUAACAAGUUUAAUUUUUUGG
AUGGAUUAUUGAUAGAUUUCAUUCUGGAUUAUUGAUAGAUUUCUCCUGCACCUCUCUCUU
UUUUUUUUUUUAAUCGAUUCUGUGAUUCAAAGUCUGUGAAGUCGGUUUGAGAAUUUUGAC
GCUAUUUCUCGAAACUCAUGUUUGUUGAUGUGCCUAGAAACCUAAUUUUCAUACUCUGGU
UGAAACCUAAUUUUUUGACUCGGGUACUGGUUUGUUUAUACUAAGCUUGUUAUUUUAUUA
AUAGAAUCACUUAACUUGGAUUACUGUGUUCGUCGUCGUCAUGUAAUGUUGCUCGAUCUG
AUUGUUUGCAAGCAUGUUGAUACUUGGUUUCUUCGACUGAACUUCAAUAACCUUGAUUGA
UUGCCUACCUCUCUCAGCUGACUUGAUUCUGGACAGACUUUUAUGUGUUAAACAUGGUAA
CUUGAGUUUACAUUUUGGUGUAACACUUUCAAAGCCAGAUAUUCUUGCUAAUUGCCUAUU
GAUUUCAAGCUAGGUUGAUUAGCAAUUGAGAUGAAUUUGAUGAUUGAUUAGAGGUGCUAA
AUCUAAUUUUUGACUUGUUUUCCUUUUUACUAGUUGGAAACAAUGUUAGUAUUCGACUAA
UUAUGGCAUGUUUUGACUAGAUUAGCUAAUUAUUUGAUAAUAUGUGUUGAUUUAAAUCAU
GAGAUUAUCCUAAAUCUUUGCAUUUCUUACUGUUUUUUUUUUUAUUUGUGUUAAUUUUGU
GUCUAUUCAUUUUGGUGAACUUUGAUUCAUCCCAGCAUGCGCUCUCUAUAAAUUGAUACU
AUCUAUUAGUUAUAGUUGGAUAAACUAAUUCCAUGGUUUUGGAUAAAAUCUCAAAAUCUU
GGGAUACUUUUGAAGUCUUUUACUCUUUUCUGUUCUCGAUGAUAUAUUUAUUGAAAAUGA
AUAUUUUUUCCGCUAUUUAACUUUUACUCCUUGUAUUUAGGAAAUAGUAUGUAACGGCCU
UGCGGCUAUUUUCGUGAUUCCCGUAAUUUCUCUGUAUUUAGAAGCCUUUUUUUAACUGUC
CCAAGUCAUUGAGAUCAGUGGGUUGGUUCCCAGCAGUUUUAUAGCUUUAACUCUUUUAUA
AUUAUAACUUCUUUGGGUCUUACUCUACUUAUGCGAUUCUGAAUUGCCUUUUUUUCUAUU
UCGUUGUGUCUGUAAUAUCGAUUACAUAAAGAUUUCAUAUAUGUGUUUGUUUAAAUCAGA
UUUUGAUUUCGGACGAGUGAUUGAUAAAUCGGCUAAUAGGCCGAUAAAUUAGCCAGUGCC
AAUAAAUCAGUCCUUUUGGUUGAUUUAUCGGCAGUUGGUGGGUCUUUCUAUACUUGUCUU
CAACAUGAAAUUUCUCAUUUUUGCUUUCAAACUUAAGUAAUUCUCUUGAUAUUUCAGUGU
UAAUUAACGGUAGUUGAAGGACUGUAUUCAAGUUUUAAUCCGUAUAAAUUCAUUGUGGUG
GCUUUAUGAUCAUUUCUGCCGGCUUUGUGCAAAUUCGAGUUUCAUACAUAAUUUUUUAAA
CAUUUAGGCACGUUUUUUGAGGAUAUCGCUGGAAUUCAAGUAGGCUAUGGUUUAUAUCUU
UAGAUUGGGUAUUAGUGUAGUAUAUGUAUGGUGAAGCACUAGCUCGAGGUUGAGAUUACU
AGUCGGACCCUGAUAUGCUUAUUUUUUUGGUCUAUAACCUACUUAGCUGACUUGUAGCUU
UAGUUACUUGGGGCUAUGUUUCCCUACUUGCGUGUUAUACGUUGACUCUGGAACAUAGUA
UGAUUAGGGAGGAGUAUAACUUUUAGUGCGUAUUAUUUAUAUGUUGACUCUAGAACAAAG
AUUGGUUGAGGGGAGAUUAUACUAGUGAAUAGAGCUUAGUAGUUAUCUCUCGAUUUAAGC
UUGUGACAUUAGAGCUGAUGAUUUUGUGGCCCUUGGAACGAUUCAAAGUUAAAAGCCACA
CUUUAUUCAUGUAUCAUAUGAUCUAAGUGAUUAAUGAUAAGACAAGUCAAGCCAAAAUUU
GACAAAAGUUUUUAAAUUUUUAUUUUGUUGUCCUAUUUCAGAUAGUGCCAAUUUUUUUUC
AAAUACUCGUUGGUAAGGGUCUCGAAGAAGCUAAAGAAGUGAUGAAAAAUAUCAAGUUGU
AAAAAGGGUACAAGAUUAGGAAAUGAUGUUGCAAAUCAUUCCUGGCCCGUGGAUGACUUC
CCUUGCUAAUAGUAGGAUUUCUCCUUUAUGUUAUCCUUCUCCUUAGUUUAAUUUUGUUAU
UUUUUUCGUUCUGUAAAAGAUUGUCGUAUUUCUCUCGUUAUUAAUGAAAGUCAUGAUUUU
UUGUCUUACUCGAAACUUUAAAUGUCAAAAUGUGGC
2. MFE structure-
(((((......((((.(((((((..................................((((.....((((....))))...))))..(((((((........................))))))).........(((.((((...........((((.((.((.(((.....((((((((((((((.(((((........
.(((..((.......)).))).........)))))..)))))).))).)))))......))).)).)).))))....(((((((((.(((.(((((......((..(((......))).)).................((((((((..((......(((.((((((((((((.((((((((((((((((........(((
(..((((........)))).)))).......((((((((.....((((((((.((..((((..............................))))...)).))))))))))))))))..))))))((((((((............))))))))...((((((....))))))....((((......)))).(((((.(((
(((((.......)))))))).)))))..))))))))))..(((((...((.((((((....)))))).))....)))))..((((((((((((((.((((((((((...((((((((((((.......((((..((((((...((((((.....))))))...))))))..)))).......((((((...((((((.((
(((((.....((((((....((((((.....))))))......((((((((((....)))))).))))(((((..((.......(((((((((((((((((....)))..)))).))))))))))......))..)))))...((((..((((((.((((((.......(((((((((((.((............)))))
))).))))).....((..((((((((...........(((((((((....((((....))))...)))))))))((((((((((.((.(((((..(((((((.....(((((((.............((((((((((..........(((.((((((((......))))..)))).)))..........)))))))))).
...((..(((((((((.(((.(((...((((((.((((((((((...))).))))))).))))))......((((.........))))..((((((((((((.....)))))))....))))).))).)))))))..)))))..)).((((.......)))))))))))))))).))..))).)).)).)))))))))).
))))))))..))...))))))..))))))..)))))))))))))))))))))))..(((((((.((..(((((.((..((((((...(((.....)))...))))))...))))))).)).)).)))))...(((((((((((((((((.............(((((((.((.((..((((...(((((((...(((((.
(((.((((((..((((((.....)))))).(((((((((...((...(((.((((((..(((((((((((((((((...................))))))))))))))))).)))))).)))...))..)))))))))..((((((((((.((......((((((........)))))).......)).)))))).)))
).....((((..(((((..((((.......))))))))))))))))))).))))))))...)))))))........(((((....)).)))...))))..)))).))))))).............))))).)))).....))).)))))..........))))))...(((....))).)))))))))))).........
.....(((.((.((((((.((((((.((((((((.((((.(((((((((..((((....)))).....)))))...............)))).)))).))))...)))).))))))))))))))))))))))))....))).))))....)))))))))).)))))))))))))))...))..)))))))).........
.((((........))))..)))))))).)))))))))....((((((....((((((((.....(((....)))))))))))....)))))).)))).)))...((((((..((..(((........)))..))))))))...........((((((.(((((.....))))))))))).(((((((((((......)))
)))(((((((....)))))))......)))))(((((...)))))(((((((................)))))))))))))).))))((((((((((((.(((((........(((((((.....((((.......((((((.((((.......))))))))))))))....)))))))..((((((...))))))....
..........))))))))))..))))))).((((((.(((((......(((.((((.(((..(((((....((.(((((...))))).))..)))))..)))...))))..))).....))))).)))))))))))
You can download the minimum free energy (MFE) structure in
II. Results for thermodynamic ensemble prediction
1. The free energy of the thermodynamic ensemble is [-647.77] kcal/mol.
2. The frequency of mfe structure in ensemble 2.92327e-38.
3. The ensemble diversity 738.92.
4. You may look at the dot plot containing the base pair probabilities [below]
III. Graphical output
1. A note on base-pairing probabilities:--
The structure below is colored by base-pairing probabilities. For unpaired regions the color denotes the probability of being unpaired. The reliability information of RNA secondary structure
plot in the form of color annotation, Positional entropy ranging from red (low entropy, well-defined) via green to blue and
violet (high entropy, ill-defined).
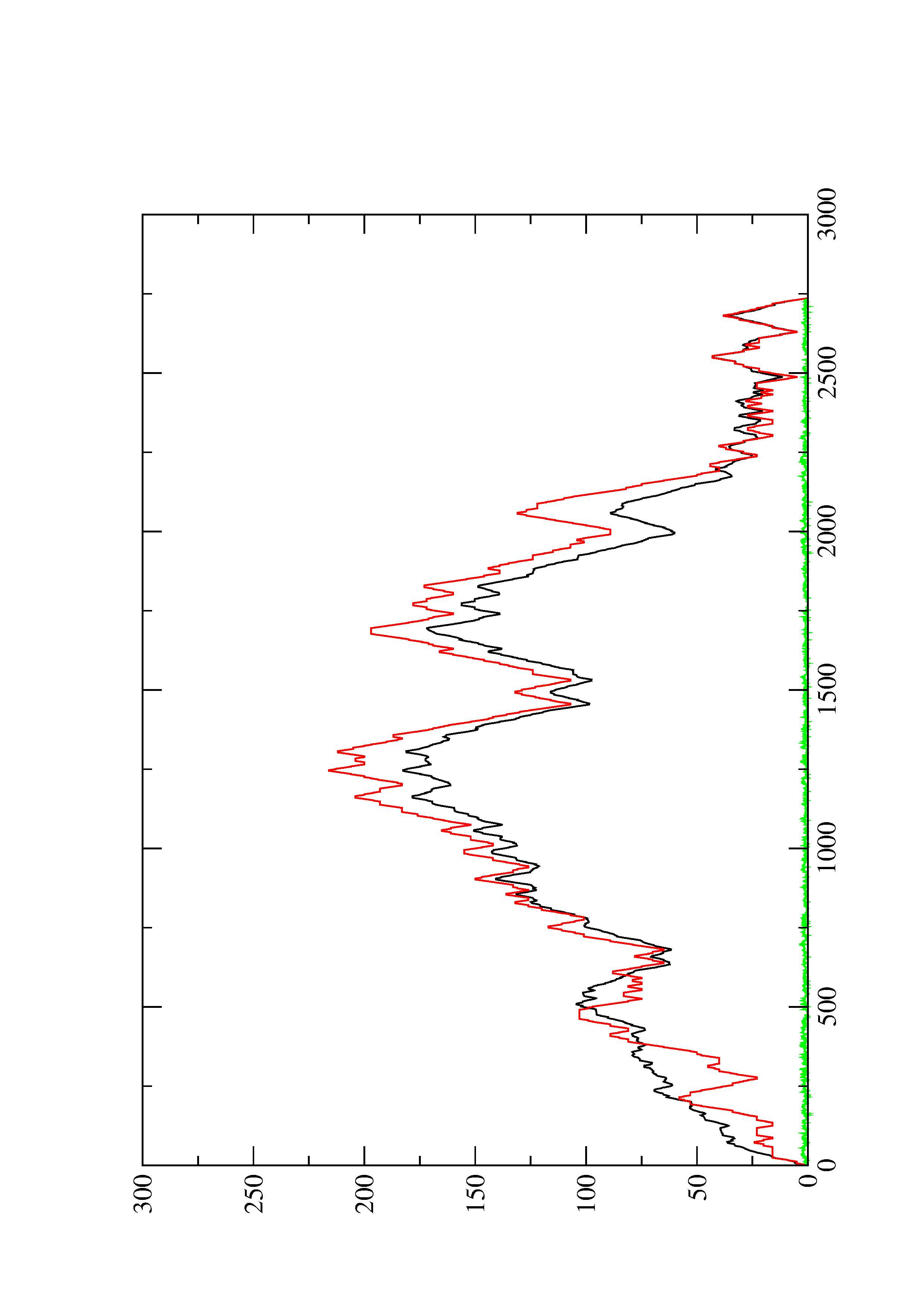
2. Here you find a mountain plot representation of the MFE structure, the thermodynamic ensemble of RNA structures, and the centroid structure. Additionally we present the positional entropy for each position. The resulting plot shows three curves, two mountain plots derived from the MFE structure (red) and the pairing probabilities (black) and a positional entropy curve (green). Well-defined regions are identified by low entropy.