lncRNA Structure Prediction
Species Name : Withania somnifera Sequence Id : GBHJ01016978.1
I. Result for minimum free energy prediction
The optimal secondary structure in dot-bracket notation with a minimum free energy of (-625.50) is given below.
1. Sequence-
CUUUUUUUUUUUUUUUUUGUGAUUUGGAGGUGUUCACUAAUUUCCUCUACUUAUUUUGUU
GCAUGCUAAGUUAUAGACCAUCUUUUAUUUUCAAAAAACUCGGUAUUAAAAGUUGAGAAU
GUUCACAUCCCUUUACUUAUCCUAGUUGCACCUCAGUGGCCCAUUUAGUUCAAUCUUUUC
UAGACUAUGUAAUGAUCUACGAAGAGUUCUCUUUUAGAAUAUAAUUGUCUAUGUGUGUUU
AUUUAGCAUGCUAUCUUUAUUCUGUUCCUGUGUAUAAAUAUUUUUGUGUGUAGUCACCUU
GAAAUGAUUGACCUAAUUGCACUAUUUGAUCUCAUAGUAAUAUGCUAGUGUUAUUUAAUA
UAGAUUCCUUAUUUGCAAGAUAAUAAGACUUAUGGUUAAAUAAAUAUUAGUACCAAUCAA
CUAGUUUACAAUUGCCUGCCUGCUUAUAUACUAGAUGACCUUGUGUGAAUCAUGCACACA
CUUGAAAUGGUAUUUAGAUUUAGUUUAUCAUGAAAUUGAGUCCCUUAUGCAGUUGAUUUC
CUGUAAAUCCUUUUGCCUAAUUACAUUAAACUUUCUGCAGUCUCUGCCUUAAGUUGUCAU
UUUAUUUUGUUCUUGACCAUGCAUAUAACUAUGUAAACUAGUAUUUGAAAGUUUGGUCUU
UCUUUAUCAUAUUAAGUCUACUCAUGUAAAUGUGAGUUAGAACCCCAUGUUAGUUGUUGU
GAUUCUGUUGAUCCAUAUACUAUCCUAUACCACCUUUUCCCAUAAAGUGAUAUAUGUGUA
UGCUAAGUGUGAUUGAAUCCAGCCCACCUAUAUCAUAUCAAUGUAAUAAUCAUGACUUCU
CCUUGGUUUAGCUAAACAUGUUUGUCUAGUUGCAUCCCUUGUGUAUGUUUGUAUCCCCUG
UAAGUUGUAUCAAAUAAAUCUACUUUGCAAAUCUUAUGGUUCUGUCUAAAAUUCGAAUAG
GUCUACUUGCUAAGCAUUACAACUUAAUAGAGUAAAGAAUAUCCUCCCUAUCUGGCUACU
UUGAAUAGUCUUAUAUAGCUUCCCCGUCAUUUUAAUGAUACUAUCUGACCUCCUCAAUCU
GGGUUAACUGUAUGAGAAUCUUGAAGCAUUAAUACUCCAAUGGAUAACCAUUUAUAUAAA
UUCUACCCACCUGUUUAUAUUUAUCUUUUAGCCUGUCUUAACUGGAUUAAUGGUUAGUUG
GGUAGUUCUUUGAUUUAUAAUCACUUAUAUGACCUUGUUCAUGCCAUAACAAUCUAGGCU
AUCUAAAUUUGAAUCUAAAGAUAGAUAAUACUAGGUCUAAAGGCUGCAUAUAACCUACUA
UACUUCCAGUUGUAUUCAUGACCGAUUUGUUAGUUUGUUUGCUCGUGUUAUCUAUACCUA
UGCUAGUUUGGUUAAUGAGAUCCUUACCCCCUGUUAAUAUCCUUUGUCUUACAACUUGAU
GAAUAUAUCUGAGUGUGAUUAAAUAAUGAUUUAUUGAAUGUUCCCUUAUCAGUUAGGAGC
CCCUUUUAUGUUAAUUAAGUAUGUACUUGACCUUAUCCUUAAUAUGUGUAACAAAUCUUG
AUUAGUGUCCUUGAUGUUAUUUUCAUAAACUUGUGAACUAAAUAACUCUGUAUGGUAAUU
AGGAGAUGUUUACUGGUCUUAUUCUUCACCAUCUGUGAUUAGAAAACUAGGGUGUCAUGU
AUGCUCUAAUGUGUUACAAUAUGAUUUGCCUACUUGCUUAGAUCUAUCAACCAAUCUGCU
GCAUACUUGUUUGAUGGAUUGAAGCUGACCAUUGCCCCUGUAUAAUUGCUAUUAUAGUUG
CUCAACUUGGUACCAUGUACUGCUUGCUCAACUAUUGCAAUUCAAUUAAAAGAAAUGCAU
UUAUAUAGGUUUAGAUACUGCCCUCAUGUUCAUAUAAUUUAACUGUCCUUAGAAUACUGU
UCAUCCUUAGCUUAUUCACAAUGCACAUGCCCUCGAUGGUUGUUUGAUUUAUAGCCUGGU
AUAAGCAUAUUGUGGAACUACUGUGCCUUAAAUGAAUGUUUUGAGUCUACGACAUUAAUU
AAAUCUUCUUAGUAACUACCUCCCUUCUGUCACGAUAUCUUAUCAUAUUUUCUCUUAGAC
UUCACUAAUAGGUCUGUCUUUCAUUAAUUGUAAUCACCCUAAUCAAAUUUAGAUGGGUAU
UGUAGUAAUUGUUUUCAUUAAUUCUUUGAUUCACAAGUAUUUGAUAUUUGGUUAGAUUGG
UUUCAUUUCCUUUAAUGUGCCAAAGGUUGUCUUAGUCUAUCAUGGAUGAGUGCAUGCCAU
GUAUGCAUUCUUGCUGCCCUUCUCUGUUUGAAUUAUAUGUGUCACGCCUUCCUCUGGUGC
AAUGCUUGAUCCCCUAAUCCCAACCCUGCAUAUUGUUCCUAACUAGCCCAUAAUAACUUU
CCUUUUACCUCCAACAUGAUUAAAAGAUAUUGUAGGUGUCUCAUCCUACUCAUUCAUGCA
AAUAUUGAACCUUUAAUAUGGCGUCAUCACUAAUUUCUUAUUGUCUAUAUUUGUACAGCA
UGACAAUAAAUCAAUAAACUAUUGGUAACCGUUACUCUUUAUGAUUGUGCUACUUGCUAA
UAUGAUUCUUCAUCCUAGUUGACAUGAUAUCUUACAUCAAUCCUGCUGUACGAAAUGCUU
GUUUAUAUGGAGCAUAGAUCUAUCUUUAGCCAACUAUUUUGAUACUAUAUGAUUCCUGUA
UUAUCUUUUAUCCCCAUGCCCCGUAAUGUGGUCUUGAUUUUGACAGAAUGGGCUUAUGGA
UCAAUGUGUUUCUUUAUUGACCCAUGUGUAUUGUCAUUUGGGCUUUGUGUUGCAUACUUU
UUGACUGAUUUAUAUGCGCUGGGCUUGGCCCAUCACUUUUAUGUAGUUAUUAUCAUAUAU
AUCACCAAUGUCUUAAAUAAGUAUUUCUUUAUUUCGUAUCCUUGUUUCAAUUAUUUCUUA
UGUGAUUUCUAAUUUAUUUACUUCGUCCUUUUUUUUCAUGUCCUUGGUGAACUAUCUCAC
GUUCCAUUUGUUUCAAACCCAUUUGCAUGCAUCUUGGAAGGCUGGGCCCGGACGGCUCGA
GAAACUCUUUUUUUCUCUCUCCCUUUCUAUAUUCUUGCACCUUUAUUUUUCUUGUACGAA
AUUAUUGUGUAUCUCGGUGUAGGAGAACUCAAUCUCUUGGAAUUUUUUAAAUAAAAAUCA
UUGAUUUCUUCAAAAAAAA
2. MFE structure-
..........(((((((((.......((((((((((((((...............((((((((((((....(((((((.((((((((...(((........((((...(((((((.(((((((((.......(((.((((..(((((((((.(((((((..((.((((.((((((.(((((((((...((.(((.((((.
(((((....))))))))).))).)).))))).(((((((.....)))))))..................(((..((.((((....)))).))..)))...)))).)))))).)))).))..)))...((((((((.((.((((((((((((((((((...(((((((((((((........))))))..(((((((....
(((((...(((((((.(((.(((((........((......))......))))).)))...((((((.......)))))).......)))))))...)))))....)))))))..(((((...((((((((((((((.....(((((....)))))............(((((...((((...))))...)))))((((.
.............))))...((((((((((((.((((.((((....((.((...((...((((((((..((((((((((((((((......))))).))))...(((....((..(((((((((..((((((...((((......)))).................((((((((.(((...(((..((........))..
))).))).)))))))))))))).....)))))))))..))...)))..((((((.(((....))).))))))......(((((.(((((((((....((((((((((((................)))))..))))))).((((((........)).))))......))).))))))))))))))))))..)))))))).
..))...)).))..)))).)))).)))))..))))).))......(((((....)))))............((.......))))))))))))))))...)))))..............(((((....)))))..(((((((((((((((.(((....((((.((((.((((......))))..)))).))))..))).))
)))))......)))))))).........((((......))))(((...........)))...........)))))))....))))))))))))((((....)))))))))).))..((((.(((..((...((((..((((((((.................))).)))))..))))...))..))).))))..))))))
))....................((((((.........)))))).....))))))))))))).))))))).....))))))))).......((((((((((......(((((..((((((((((((((.((...........(((((((((((((((...((.((((((((.(((((......(((((....)))))((((
((((((.((...(((((...((((((((...((((((((.(((.....((((((((((((((.....((((((((.....))))))))..((((((((((((((...........((((.((((((((((.(((............))).)))))))))).))))((((.......(((((((((........(((((((
..(((..((((((...)))))).)))..)))))))((((.(((........))).)))).)))))))))..(((.(((((..................((((.((((((((((((...((((((.....((.((((((((((((..(((((......((((((.......)))))).)))))..))))..))))))....
.)).)).....)))))))))))))).....))))))))...........))))).)))..........)))).((((...))))...........(((((((.......)))))))....))))..))))))).))).)))))))....)))))))((((((.((((.((((..(((.........)))...)))).)))
)))))))..))).))))))))..))))))))......))))).))))))).))))).))))).))))((((((((((...))))))))))..))))....))...)))))..))))))))))..........)).))))).))))))))).................)))))..)))))))))).........)))))))
.....)))).......))).))))))))...(((((........)))))......((((..(((((((....))))))).))))..................))))))).......)))))).))))))(((((((...)))))))...(((........)))............((((..((((....))))..)))).
(((((((.....((((.((....)).)))).((((((((((((((((((.((((((((.(((....)))..........((((((............))))))..........(((((.(((((..(((.............)))..)))))..))))).((((((........))))))((((.(((......))).))
)).)))))))).))))......(((((((.((((((...(((((...)))))..(((.......)))......)))))).)))).....))).))))))))))))))........(((.....(((..((((((.......))))))...))).....)))..............))))))).))))))))).)))))..
.(((((..(((..((((....))))...)))...)))))....(((((.....)))))((((((.......)))))).........(((((((..(((((....(((((........)))))....)))))....)))))))(((((.(((((......((.((((......)))).))))))).))))))))))))))
You can download the minimum free energy (MFE) structure in
II. Results for thermodynamic ensemble prediction
1. The free energy of the thermodynamic ensemble is [-686.70] kcal/mol.
2. The frequency of mfe structure in ensemble 7.53304e-44.
3. The ensemble diversity 883.17.
4. You may look at the dot plot containing the base pair probabilities [below]
III. Graphical output
1. A note on base-pairing probabilities:--
The structure below is colored by base-pairing probabilities. For unpaired regions the color denotes the probability of being unpaired. The reliability information of RNA secondary structure
plot in the form of color annotation, Positional entropy ranging from red (low entropy, well-defined) via green to blue and
violet (high entropy, ill-defined).
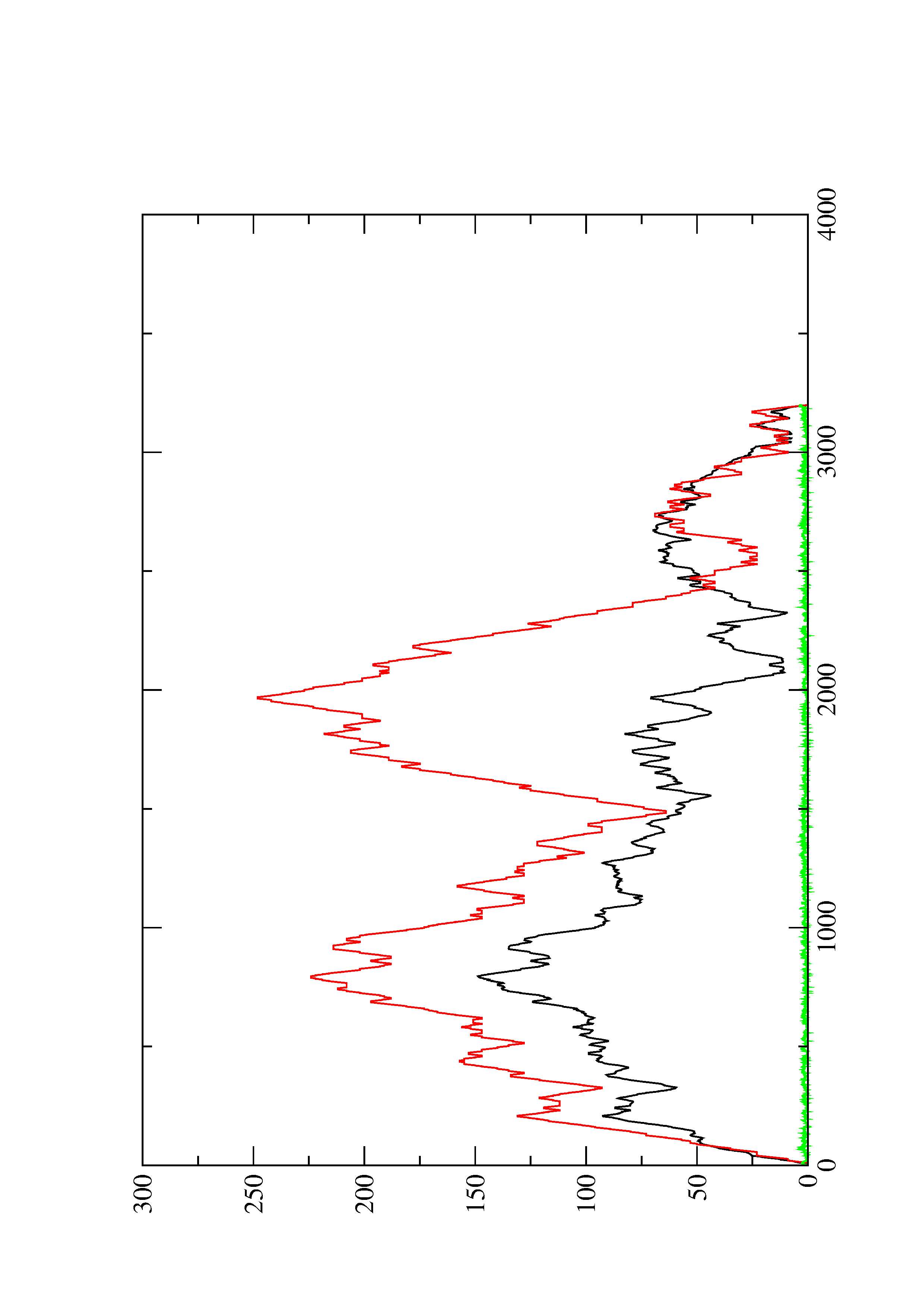
2. Here you find a mountain plot representation of the MFE structure, the thermodynamic ensemble of RNA structures, and the centroid structure. Additionally we present the positional entropy for each position. The resulting plot shows three curves, two mountain plots derived from the MFE structure (red) and the pairing probabilities (black) and a positional entropy curve (green). Well-defined regions are identified by low entropy.