lncRNA Structure Prediction
Species Name : Withania somnifera Sequence Id : GBHJ01017608.1
I. Result for minimum free energy prediction
The optimal secondary structure in dot-bracket notation with a minimum free energy of (-642.40) is given below.
1. Sequence-
AUGACGAUUUCAUUAUUUAUAAAAUCAAGUGUUGGGUUCUCGUACAUUGAUCUACACCUU
CGAUCCAUGAACAACUAUACUAACAACACAUAAAUCUAAGUACAAAGAAGAACAAUGCAU
UUGAAACUUGGGCCAUCCAGGCCUAAUACUCGCGAGGCCAACUCUGGUAGGCUUUGGACC
CAUCAAACACGUAGAAAGAUUGCCAAGGAUAUGCAAAAAAACAAGUGAAUCCAUUAGAAA
CACAUAAAAUAUAAUAAAUAAAAAUAGGAAUAACCAACCAAACAAUAUAUUAAUGAAACA
AUGGACCAACUAGAGAGCAAUAAAUUACACCACUAUUAAGGCCCAACAUAGAAUGCAAGA
AUUCGUAAGACAAUUUAAUGGCUUACUUAGUUUCCCCCUCGGAGCUCAAACAUAAGCUAC
GAAAAUACACAUGUUUUAUUGAGCUUUAACAGGCCUAAAUUAAUUAUUUCAAACUAAAAU
UGAAUAUUAUUGAAGAAAGUGGAAAGCAUGCAUAUAUUCAUAGCAACAAAUAGGAAUGUA
AGAGACAAAUAGAUAUGACGCUCAAUCAUAUGACAACGCAGGAAACGGAAGCAAGUUGUU
CAAAUAAAAUAUGCAUUCGUUACCACCAAUGUCAUGAUGCAGGAAAUAAAUAAAAAUGAU
CGGGUUUAUCUAUUUUUAACAAGGAACGUGCAAAGAUAGCAUCACAAGUAAUAGGCUAAG
GGCAGGAUGUCAUUUCAACCACGUCAAUAGCACAGAAUCAAGAAAAGAUGUACAAGAGGA
GAUUAUAGAGAUGCAGGGUGAGCAUGCUUACUCACUUAAAGGGGAGGUUUAUAAUGAAUG
CGGGAAGCAAAUGACUAAUAGCAACAUUAACGAUCACUUUUGACACGAGAGGGAGAAAGG
UGUGCAUAUAAGGUCAUAUGGUUUUACUAGGUAGGCAUGGUUAACUAUGAUAUUCAGGGA
GACCCCAUAGACUCAACAAAGGAAGAAAAUAAGCAGUAAGAUAGAAAGUUAUCAGGAUAC
AUAGAAGUUAAUCAUGAACUUAUUAAAGCACCUUAAAACAACAGGAUGGACAGGUCAUGC
UAUACUCCCGAUGACUAUAUAUUAUAUUUUAUCCAUACAUAAAGGUGAGAUAACUUAGUA
UAUAUAACAAGUAUGUAUAAUUAAAUGAUGUAUUUGACAGUUAUGCAUUUGAGACCCUAA
GAGUAUUUGACCAAUUAGUGCAUAAUUGAUCAAGCAUGGAUUAUUACAAUCCUCCUUAUC
UUACAAAUGAAGGGACAAUACUAGCAUGGUUCCAUACACACAGGUGAGAUGAUCUACUAU
AUAUAACAAGUAAGUAUACUUAAAUGUUGUAUUUGACAGUUAUGUAUUAGAGACCCUAGG
AGCUCUUUAACCAAUUAGUGCAUAAUUGAUCAAGCAUGGGUUAUUGAUACAAUUCACGAG
UUAACACGAAAUCAGGGCAGGGGUAAAGUAAAGAAAACAUUAUAUUGCAAACAAAGAGCA
CUUAAAACAGUAUAUAAACUUAGCAUAGAAAUCAACCUUUGGAGGCAAAUAUGAGCUAAA
GAGAUUCUUUGAGGAAUUGAAUUUCCCCAUGUCCCUCAACCCCUAGAGGAUUCAUACCCU
UGGCUUCCUAGGAAAAGGUUUAGACUCUAUACUAAACCUUAUACCUCUCCUUAGGCACUC
UCAAAAGGUGCAAGCUUUUAGAUAAGUAAAAAACUUCAAUUCUUAUCACAGGCUACCAUG
UGAGUCAACUUAAAACAUUUUGAGGCUAUAAAAGACUCUCACUGAACACCAAACAUAAAG
GGACAAUUCUAAAACAAGAGAGUAUGGGGUGGGUUGAGGGUGUUUUAUUGUAGAACUAGA
GAUAUCUAAUUGAUUAUAUAAAAAUCAGUUAGAAAUGGGUGUAAAAUGGCCUAGGAUAGC
CUCCCCUACCUCUUAAUUCCAUACGAAACCUUUGAAAGUUCUAUUUACCCUUUAAAAGAG
ACAUCUUUUAAAAUAGAAGAAAACUUUGGCAAAUAGAACUUGAAAAAAACCAUUUUGACA
AUCAACUAAAUUGGUAGAACCAAAGACAUCAAAGACUCAUGACUUGGACCAGCAAUAAAU
CAGGCAGCAGAACCAGUAUUGGACGAGCAACAAUACUAAACGAAGGACUACAUGACAAAU
AACCAAUAAGCAAUGAUCAUAACACACACAACAACUAAAUCACCUUAAAGACCACAUAGA
GAACCUUUAUAGUGACCAAAGUUUUAGCAUAAACAACAAAGAAACAAGUCUAUCACAGAA
AAUCAAAUAGACCUCUAAACCUCACAUAAUUAAACUUUAGCACGAAAUCAAAACUAGAGA
UACGAACUAGUCAAUGAACACCUCGUCAUUAUAGGACAUUCUGAAAUACCACAUUAUAUC
AAAAUAACAAUAAAUGGACUAGAAUAGACAAUAAAAAAGAAAGGGACAAAUUAUACCUUU
UAAGGUGCAGGAACACAACUUAGACCGAAAAAUAGACCUGAAAAACACUUGAAAUCUAUU
GAACUCGCUAGUCUACAAAAUCCCCUUAAAUGAGUAUACGGAAAGCCCCUUUAAGAAAUA
CUCAAAGGCAUUGACGCUCACUCUAAUUUUACCUUGACCUAAUACUCUCAAAGGUGCUAA
AAAAUGUUUAGAGUUAUAUGUACUCUUUGUCUUAACAGAUAAUCUUAGCAGUAAAUCUAA
ACCAUAGUCAGCUCUCACCUUGAAAUAUUAAACUACAUGGACACUAAACUAAAAAACUCA
GAAUCUUACUUUUUAGCCUUAUUAAAACCCUAAUAGAAAACCCUUUCUCUCACUUUUCUC
UCAAGGGGAAAAGGGGGAGGAUUUAUAUAUAGGGGAAUGAAAUUAUUCCCCAAGAUUAUC
AAGAAAAGGGGGCGGGACAUCAAAAUCCAAUCUCAAUACUAAAUUCGAAUUUUGAGAAAA
UCUCAUCCCUUCAAGGGACUUGCAUUUUAAAAAAACAUUUAUUAGAGCCAAGGAAAAUGA
UCAAGAGUCAAUCAUUGGCUCACAGGCAUGCCAAAAUCAUACAAGAUAAUGAGAGAAUUC
CCAAAACCUCAAGACACAAAAUCAUGCCCUCCCAACGAGAAGGAUGAAGGUACAAAAAUA
CAUAGAAACACAUAAAAUAUAAUAAAUAAAAAUA
2. MFE structure-
.......(((((((((((......((((((((...(((((.((.(((.((((.........)))).))).))......................(((........))))))))...))))))))...(((((((.....(((((..........)))))..(((((((.((((((((....(((.............)))
..))))))................(((..((.....))..))).......................((.....))..(((.....................))).)).)))))))........................)))))))..(((((((((((.....(((((.((......)).)))))...(((((((((((
(.((((........)))).))).......(((((((.(((...((((.(((..((((..(((((...(((((.......)))))....)))))............(((((........))).))......))))..))).))))...))))))))))....((...(((((((((.....((.((((((.(((.(((...
.......))).))).)))))).)).....)))))).)))...))...........((((((((((((.(((((((((((.((((...((((.......))))............((((..(((.((.((......)).))..)))..))))...............(((((.((((..((((.....(((((..((((((
(((................(((((((((((((..(((.(((.....))).((((((.((((((.((((.....((.((((((...)))))).))....))))))).)))..))))))...((((((.(((((...((((((((((((((.(((((.(((....))).................................(
((((....))))).))))))))...))))))))))).))))).))))))...............(((((((.(((..((((.....((.(((...(((((((..((..((((((((........))).)))))..))..))))))).(((((((((...........))))))))).((((((((((((.........((
((((.((((.....((((((((((((((.((((....(((((.....)))))...(((((((((...((..(((((.((......)).))))).....))...)))))))))........((((((((..((((....))))..))))))))))))))))))))))))))......)))).)))))).........))))
))))))))..((((((((((((.(((((..........((((....((((((..((.(((((((...(((..(((....(((((((((...(((((..............((........))..............)))))...)).)))))))..((((((...)))))).((((......))))...(((((((....
.(((((.(((.(((......))).))))))))..(((((((((........)))))))))..............(((((.......))))).(((((...((((((...............))))))..)))))..((((.(((((..((((((....)))))).........))))).)).))..............))
)))))(((((........)))))..((((((((..((((.(((((...................(((((((((((.......)))))))))))..(((((........)))))))))).)))).))))))))....)))..)))...))))))).(((((((((((.((.(((((((((....)))))))))...((((.
...)))))).)))))))))))........))))))))......))))...(((((...)))))....))))).)))))))).))))............))).)).))))..)))(((((((.........)))))))..............((((((................)).))))...................(
(((..(((((..............)))))..)))).....((((......)))).............)))))))........))).))))))))))...)))............((((((.............))))))..........(((((((..(((((.((((((.(((..((((.(((.(((......))).))
)...........))))..))).))).))).........((((((...........)))))).)))))..(((........((((((((..((((((....((......))....))))))....)))...))))).....)))((((...(((((((....((((...)))).....)))))))))))))))))))))))
)))).)))))(((((...............)))))(((.......(((((((.((((.(((.....((((((....)))))).....))))))).))))))).......)))))))..))))..)))))...................(((((((..............))))))))))).))))).....)))))))))
))).(((((((.(((((((((...))))))))).)))))))..........((((((((....))))))))..)))))).........)))).)))))............((((((...............))))))........(((((...))))))))))))))))...............(((((((.((...(((
(.....)))).)).)))))))...((....))...........)))))))))))...........((((............((((.(((((.....)))..))))))))))...........................................
You can download the minimum free energy (MFE) structure in
II. Results for thermodynamic ensemble prediction
1. The free energy of the thermodynamic ensemble is [-693.23] kcal/mol.
2. The frequency of mfe structure in ensemble 1.52474e-36.
3. The ensemble diversity 641.82.
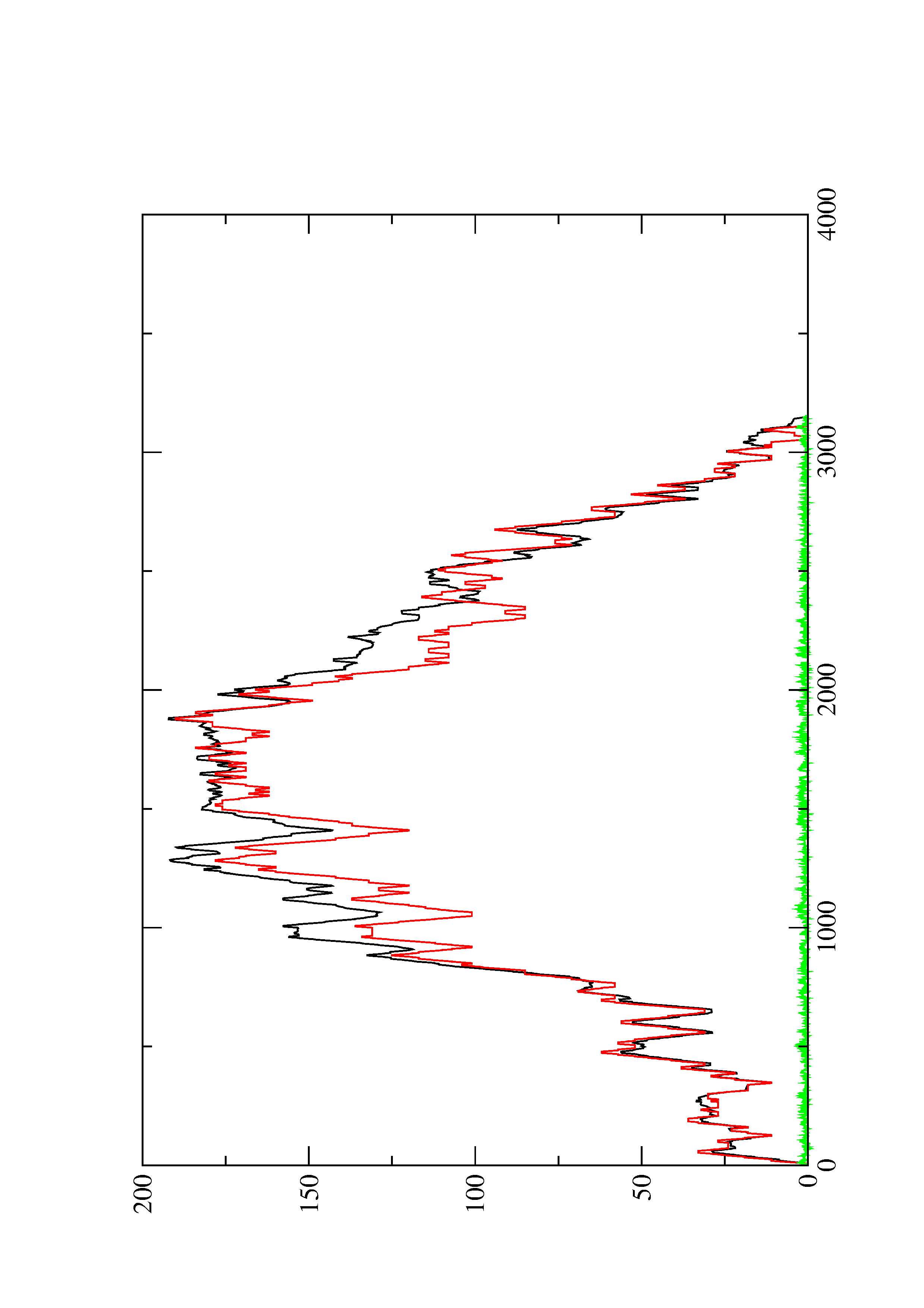
4. You may look at the dot plot containing the base pair probabilities [below]
III. Graphical output
1. A note on base-pairing probabilities:--
The structure below is colored by base-pairing probabilities. For unpaired regions the color denotes the probability of being unpaired. The reliability information of RNA secondary structure
plot in the form of color annotation, Positional entropy ranging from red (low entropy, well-defined) via green to blue and
violet (high entropy, ill-defined).
2. Here you find a mountain plot representation of the MFE structure, the thermodynamic ensemble of RNA structures, and the centroid structure. Additionally we present the positional entropy for each position. The resulting plot shows three curves, two mountain plots derived from the MFE structure (red) and the pairing probabilities (black) and a positional entropy curve (green). Well-defined regions are identified by low entropy.