lncRNA Structure Prediction
Species Name : Withania somnifera Sequence Id : GBHJ01018130.1
I. Result for minimum free energy prediction
The optimal secondary structure in dot-bracket notation with a minimum free energy of (-564.50) is given below.
1. Sequence-
CGCCAUGUGACUGAAUUUGAGAAAGUAGUUCAAAAUAACUCUAAAAGGGAUUCAUCCAAG
UUAAAAUGAAAUGAACAAAAUUCAAGUGUUGGUAACAAAUAUGAAAUAAAAAAUAUACUC
CAUGAGUAAAGAGAGGGAUUUUGAACUAUGAUCCAAUAGCCUUCCGUUCGUUCCUUCUUC
UUUAGUAUUGACCGCCCCAGGCUGUUGUUGCUGCGAUCUUAAAGCCGUUAGGGACUUUGA
ACCUUUAAGUUGCUGGGCUGCUGCUAUUGUGACAAAUAUUUUGAGCUUUUUGAGCUCACU
UUCUUACUAUGUUGAUGUCGGAUUUUCAGGCCUUUGGCCUUAUAUUUGCUUCCAAUCGAG
GGGCCUAGCAGAGAUGACUCUUAGAGGAGGUGGAGUCCCAUCGGACUCGGAUAAGGAACG
GUAUUCAUGCAAAAGAAAGAGGGAAAAGGAUUAGAGAGUGUAACUUUAAAAUAUCAAACA
GUGCACAGUACAUGUAAUACUUCAAAGUGGAUCUUGAUGCUUGGAUUUUCAUAUAUACAU
GUAUAAAUAAGCGAGCAAACUUGCGUAAAUAACGAAGCACCUUAACGGGAAUUUUAAGAG
UAUUUCUGAUUAGAUAUUAGCCCAAUUCCGAUAUCAAAAGGCAGCCAUACAAAAAUGAUU
AAAGACAUGGCCUACCUAUUUCCUUAUUUUUCUUGCUUUACAAAUUCAGGAGCAAAAGCU
ACCAUAUAUUCAAGAUUUUCAGCUGUACCAUCGAGUCAAACAGCCGACAUGUUAUAAAUU
AUAAAUUAAUGAUAUUUUGAAGGUCAUAAAAUAUACUGACUUGCUGGUUUCUGCGCAGUU
CAGUUCAAUUUACGAGAAGAAAGGAAGAAGAAAACAAGGAUAUAAUAAAGAGAGACAGCU
AGCCAAUUGAGACACAAAUUCAUGCUUCAAGCUACUUCUGUACACUCAAACCUGACAUGU
CUAAACAAGUAAUAAUUAGAACCAACAAAUAUGAGGAAUACAUGCUGCUACAGGGGCUUC
AAACCAACUUAAAACACACUCCAAAGGGCAUUUUUAUGACCAUUAUUAAUUCGGAAACUU
AUAAUGAUUGAGUUACUCCAACUAAUUUACACUUCAGAUUCAAUAAAUAAUCAAACAACC
CACUAAUGAAAGGAAGGACAACACCUACUAGAAUAUGAUAAGCCAAGAUUCGCCUGAAGU
AAAAGAAAGUUGACUGUGAACCCUUCCUCCUAAUAUUUUCUUAAUCUCAUUCAAUUUCCU
UAUUUCAGGGAAUCAAUUUUAACAAAGAAGACUGCUUGUAAAAAUGGAGAUUACACACAA
UCAUGACAAAGUUACAACAUACUUCACAACCAAUUUGAUAUAUGAUAAAGAUUUAGAGCG
UGCAACCCCAGGGGGUAAUGAAGAUUACGGUCCUAGUCUUAAGGAACCGGACUAGUUUUC
CCAAAGGCCAACUAUCAACUCAUUUUUCCUUUAGCAAGUCAUACAUGAUAACACAGAAAA
GAAUGGACUUCCAAAACUUGGGAAGACAACAGAUUCAGUUGUCUUAGAAAACGACGUUCU
UUAGAAAUCUACCCAUGGAGAAUUCGAAGAUGAUAAAUUACCAGAACAUUCAAACCAAAG
ACGUAUGCAAGAAUUAGCCUUGAAGAAGAUGUUUUAAUACUAAAUGACUGAAAGCAGCUU
GAACUAAUUAGGGGAGACUGGGAGAAAACGACUUCUGGAGAUAAUUUUUAAAAGAGGAAA
ACAGAGUUUCACACUACUUGUUAGCCAAACCUGAGCCCACUUUGUAGAAAAAUCAUCAUU
ACGCGUUUUUAAUAAACAUUUACUUCAUGUGCUAGGUAAUAGAAUGGUUCUAAAUCAGAA
ACAUUAAUAUACGUCCGUAUUGGUUCCACUGAAUCUGGCCAUUAACAAUACUUAGUUGAG
UAGAAAAUAGUCAACAACUAUCAUUUCUGAGACAUAUCUGAUUCUUAAAGCCACGGGCAC
ACCAAAUAAAUUUAGGAGUUAAAGUAUAAAGUCACAAGGUUAUUCAAACUUAGGCAUCAA
UCUUUCACAUUUUCAUUUUAAUGGUUAAGCUCAUUUAGAAGACCUCAAAGCCCAACGAUC
AUGAUUUUCUACAGUUUAUUUAAGCAAGGUAAAGACUUGUCGCAGCCAAACAGAUACAAA
AGGGAGCAAGCAAAAUAGAAUAAAAAACACAAUGAGUCCCACAGGCGCUCUUAUUAAUCU
UUAGGCCGAAUCAAUAGUCUUUCCCCUUAGAAGGUUCAUGGUUCGCACAAGAAUAUGUUA
CAACUGAUUUUCUUCAAUACAAUGCAGGAGACAGAUCUAACUAAUUCACAAAAUAGGAGU
UUAGAUCUAACUACUGACGAUUUAGUCAGUAGUUACUCGAAGUGUUCUUAGCCGUUUUAA
CCAUUUUACAUACUUGUUUAUAUCACACAAACAGCGAUGUCGCGAAUUCUAACAAACCGA
AAAUUAAACGAGGGAUGGCUUUCGUCACAUAUUCAUCAAUCAAAGUUAUCAUUCAUUCCA
AAUAAUCCUAGUUUCUGCUAGUCAUUCCAACUAAUUAAAAAAAAAACACUUGAACAUAUC
UAAGAAAACAGUCAAUCAUAGGAAUUCAUGAAAAUCUAAACGAGUACAUUAUAAAAAAGG
GAAAAGUGGGGCGAGUACGGACCUUCUUCGGGGCAGCGAACUAAGGGGGCGA
2. MFE structure-
((((.(((((((....(((.((..((((((((((((..((((.....(((....))).......................((((....(((....)))...))))..........(((((...)))))...))))..))))))))))))..)))))....(((..((((.(((((((((((((.((((..((((..((..
((.((....)).)).((((...(((((...((((((((...((((((((((((((((((.((...((((.(((((((((..(((((((...)))))))...(((.((.........)).))).....(((((...))))).)))))))..))))))..)).))))))))))........))))))))....)))))))).
.))).)).))))..))..))))....)))).)))).)))))))))..))))..))).(((((((((((......(((.((((.((((((.((.....(((((((..((((((((((..(((((.....((((((................(((((....)))))((((((...(((((.((((....((((((.....((
(((((......))))))).....))))))..........(((((((((......))).....(((((((.......(((.(((((.((((((((.((((.........(((((....))).))..........(((((((((((((.............)))))....(((((((.((((...)))).)))))))..)))
))))).((((....(((((...((((((....)).)))))))))....)))).......)))).))))))))...))))).))).....(((..((((.((((...(((((.((........)).)))))))))...))))...)))........)))))))...................((...........))....
...))))))..)))))))))..........................(((((....))).))...........(((((((((.......))))))..))).....))))))......((((........))))...................((.(((......))).))...))))))))))))))))))))).))))))
).........))))))))..................((((((...........(((((((......)))))))...........)))))))))).)))......))))).)))))).............))))))).((((..(((.........)))..)))).......(((((..(((....(((((((((((...(
((((((((((((((.......))).))))...)))))))).......((.....((..((((((((((((((.....((((....))))..........(((((((.(((((........))))).....(((.((((.(((((((((...(((.((((((((............)))))))))))..(((((((.....
(((((...((((((((((.((((((((......(((((..(((((.((((((((....................((((((...((....))..))).)))...(((((((.....(((.(((..((((..((((.((....(((.((((....(((.....)))..)))))))...)).))))..))))..))).)))..
...)))))))...)))))))).)))))...))))).......((((..(((.....)))..))))..))))))))....))))))....)))))))))................(((((.((.....)).)))))...))))))))))))))))...)))).)))....(((...)))...((.((.....)).)).((.
((((......(((...(((((.....))))).)))......)))).))...........((((((((((..(((((.((.....((.((((((..((((((((((((((((..((((....))))..((.(((((((..(((..(((.((.((((...((.((((((..((......((.................))..
....)))))))).)).)))))).))))))......))))))).))..))))))).)))))).))).....((((..((((...(((.((((((.((......)).)))))))))...))))..))))........)).)))).))..(((((((((((......))))))))))).)).)))))..))))))))))....
)))))))...((((((((((...............(((.(((.(((((((...................(((.(((((((((...................))))))))).))).............(((((....)))))...........................((((........))))................
))))))).)))..)))))))))))))..........))))))))))))))..))....)))))))))..))))..)))..)))))..)))).
You can download the minimum free energy (MFE) structure in
II. Results for thermodynamic ensemble prediction
1. The free energy of the thermodynamic ensemble is [-613.17] kcal/mol.
2. The frequency of mfe structure in ensemble 5.09312e-35.
3. The ensemble diversity 670.86.
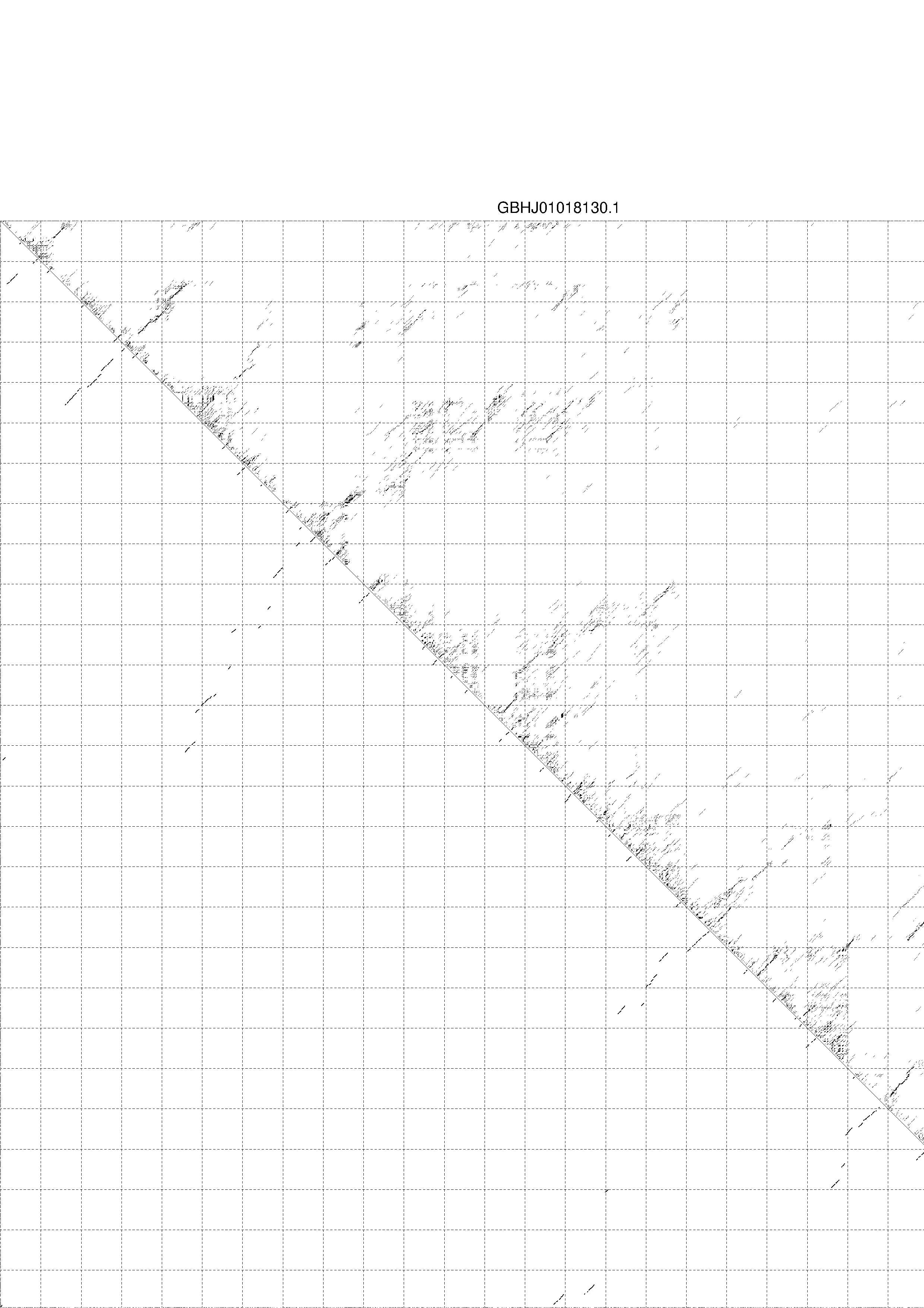
4. You may look at the dot plot containing the base pair probabilities [below]
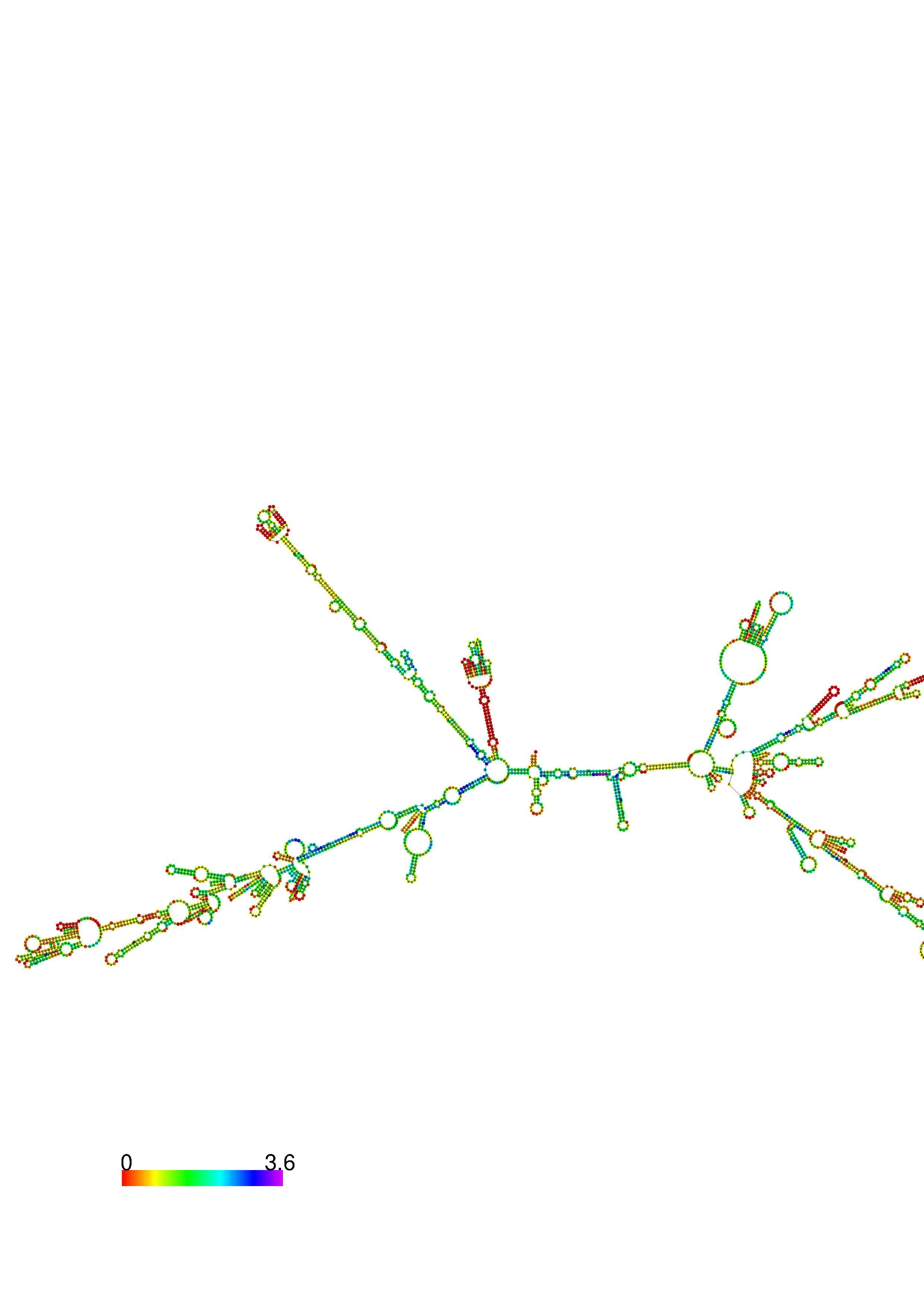
III. Graphical output
1. A note on base-pairing probabilities:--
The structure below is colored by base-pairing probabilities. For unpaired regions the color denotes the probability of being unpaired. The reliability information of RNA secondary structure
plot in the form of color annotation, Positional entropy ranging from red (low entropy, well-defined) via green to blue and
violet (high entropy, ill-defined).
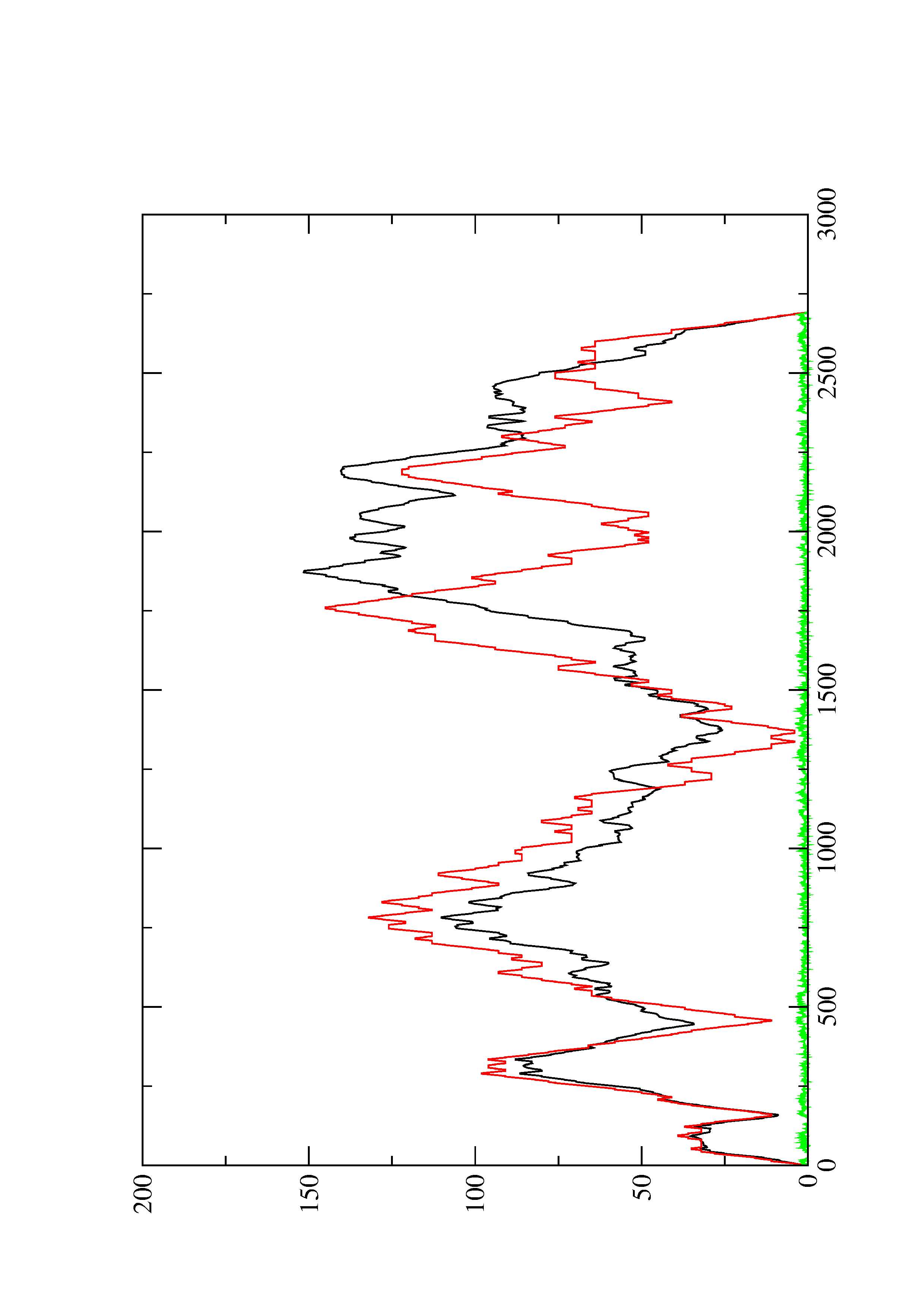
2. Here you find a mountain plot representation of the MFE structure, the thermodynamic ensemble of RNA structures, and the centroid structure. Additionally we present the positional entropy for each position. The resulting plot shows three curves, two mountain plots derived from the MFE structure (red) and the pairing probabilities (black) and a positional entropy curve (green). Well-defined regions are identified by low entropy.