lncRNA Structure Prediction
Species Name : Withania somnifera Sequence Id : GBHJ01018644.1
I. Result for minimum free energy prediction
The optimal secondary structure in dot-bracket notation with a minimum free energy of (-647.70) is given below.
1. Sequence-
CACAUGACAAAGACAAGCCCUGACACUCACCCACCCAAUGGAAUUCUCCUACAAGAUAUC
CAACAGAUGAACUUUUUGGUUGUUCAUACACCAGCAGAGCAACACAAUCCUAUAAUUGAU
AUACAGUAUUAACAAAUAUAUUGAUUCAAGAUAAGACUAGAUGCUAAAACAUAUACAUGA
CUAAGUAGCUCAGUAUUAUGAUGAACAGAUUUUUUUAAUUGGGGAACUUUGAAUAACUUU
UUUUUUUUUUCGUCAUGUCCAACAGUACGUCAGUACUAAGCUGCUCAUAUUAACAUGUAA
UGCAGAAACACAGCCACAAAAGGGUACAUAGAAUUCCUUCAUCAACUUGACCAAGCAUUC
AUAGUUCCCAACCUGUAGAGCUUAUCAAAUUCAGUCCUGCCAUAUACCGCAGGCUGCUUC
AAUUUUCCAACCUACAGAUAGAACAUAGCACUUCAGACUGUUUACUACUAACAAAACAUC
AACCGAAUACAUAUUCUCAAGACAAACCGACAACUGAGGUUCUCGUACAAUCCCAGAAAG
AGAUUGAAUAUGGUAAUUCGAGGAUGCAUCCAAACAAUGUCAAAGGAAGCACAAACAUAU
UGAUUCCUUUGGAGAUAUACGAACACGUUCUCAAAGAAAACACAUUUCUUGCUGCAAGUU
GAGGGCCCCGUUAGGCUAUUUGAGAUGAAAUUAUGAUUUGAAAUCAUGAUAUGAAAUCAU
AUGAUGACAUGAAGUUGAAGUAUUGUUUGGACAUGCAAUUUGAAAUUUUUAAGUUGUAUU
UUUUUCUUGUAAAUAUAAAGAUCUCAUAGGUUUGAAAACUAUCAAAAUUACCCCAAUAAG
AUUAAGCGCAAGUUCAUAACAAAAGUAUAAUACAUUAUUACAAAAUAUUCAAAAAAAAUA
CAACUUUCAUUGGACAAACUUCAGUUCAAUAAAAAAAAGUAAAAUUGAAUAUGAAUUGUA
GUAUAUUACUCUUUAAUAUAAUCCUAGCUCAUAGAACGGUCUCCUCACGUUGAACAUACA
UUUCUUGAGCAGAUGACGGAUAAGUCACUGUUACUUUGAGCCAUUUGUCGGUCAUAUCAU
UAGUAACUAUAUCUUUAAGUAGUAAUAAAUUUCGUUCGUAAAUGAAUUUAGUCAGAAAUA
AUAUUUAGUGAAUAUUAAAUGAUAUAGUAGUUCUGAAGUAAUAAUAAAUUUAAUGUUAGU
GAGAAUAUGAGAAAUAAAUUAUUUUAAAUGUUGAUAAGUACAAUAAUGUUGGGAUUUUUU
UUACAAAAUAUAAACUUCAGGGUUAAUUUUUGUAUCUAAGAAAAUUUCAAGUCAUGAAUC
UCAAAUCACACUUUAGGAAAAUUUAAAAGUUCAUAUCAUGAUAUGAAAUUGCAUGUCCAA
ACACUAAUUUCAUCUCAUGAUAUCAGAUUACAAUAUCAUGAUUACAAAAACCCGAUAUCA
CAUGUCCAAAUGCCUACCUCAUCGAAGUAAUGUACUACUCAGGCAGGGCUUGACCAAAAG
GAUUACUAAGGACAUGUUCUCUUACAUUCCAAAUAGAGUUUAUAUUGGAUGACUCAAAUC
UGUCUUAUUUAGCUAUAAAAUCCAUUUACCAAAAUAACCAAUAACCAGUCAACAUAACAC
AUUUUGAUAAAGUAAAGCACCAGAGAGGAAGCAAGCUACUAUAUCACAUGAAAGACUUAC
AAAUUGCAUAGUUCAGCUACCUUUUUAUUUUCUUUGGAUCGUUCAUAAUGAGUCCAUCAA
CCCCUUUUGCCACAUACCUACGAGUUUUGUUCUUUUUUAUGCAUCGCUAUUUCUGCAUCU
AUUUAAAGCUUUUGUGUCCUGCCUAAUCUCUAGAAUGAACCUAAAUUCUUCUCAAUAUGA
UUUAAACUAAUAGAAGCUAUGAAUUUUCACCAAGUUGGAAAAAAUUAAGCUAUUCGUUAA
UAGGGUGCCGCAGUGUUCUGUCCUCCAGCAUCAAAUCUCCAUAGGAUAUGGAAUGAUGCA
AUUCUUAACAAGUUUUAUACAUAGUAGAAGUAUGUGUACAGAAAACUUACCUGGACUAGC
UCAAUAAAAUUGAUGGUGAAGAAGAAAGCAUAUAGCUAAUAUGUCAACCUAAGAAGCAUA
GAACAGGUAAAAAGUGAGUCAAAAAGACCCAUGUUCUUUGCCACAUGUGGACAAAAGAAU
GGAUUGAGUUCAUCAUAACUCAAGGUAAUUGGCAAAGCAAACUGCAACUUCUUGAAAAGC
UCAUUCUCCCGCAAUUUAAUAAAACCAGAGUGCCAAUAUAUCUUCAGAAGACUUUUCCUU
CACCUUUGAAGUACACCUGAGGAGAAAAAACAACAAAAAAUAAGCACACCAAAAUAAAGC
AAAACACCUAAGAUGACUGACCCCACUUGCUCUAAACCAAGCUGUUCUUGAGGAUGAAUA
CAGCCAACGGAAACAAAAUAAUGCUAAGGAGACAAACAAGCAGUCACCACAACCUUCAGC
CAAUCACAACUAACUCCACCGGUUACCGACCACCCCCCAACAACAAGAUAACAUAUCGCC
AAAGCCUAAACAAAAAAGAAGAAAAGCCAAUCCAAUAACAACCCAUCACAGAGAAAUACA
ACUAACAACCACAAGAAAAAGGUCAUCACCCAAAGGCUAAGGUAAAGACAAGGAACAAAU
ACAACAAGAUCAUCCUCCAGGUACCUCACCUGCGUAUUUUCUCAGGCAGCAAUCUUUCUU
CGCUUCACCAAGUACCAAAUCUUGGUUAAAGGUGAUCAGUCUACUUGCUAAACAUUCCUA
AAGUUCCCAAAACUAGUUCAUCCUUUUAACUGGUUACAAUAUUACUUCCUCCACUUUCCC
UACCAAUAGAUACCCCUAAUUUACUGAUAUUAAUGACAUAAUUGUGUGCACACUUCAUAA
UCCUGAAGCAGUCAUGAUGCAUCAUGAAUGCAGUGUAAAUCCAUAACACAUACAACAAUA
ACAUAACCAGUGUCAAACCGACAAGUGGGGUCUGGGGAGGGUAGGAUGAACGCAGACCUC
ACCCCUGCCUUCCGUGGGAUAGAGAGGUUGUUUCCGAUAGAUCCAAUGACAAACACAUAA
CACAAACAAAUACAAGCCUCCUAUGUGCUAAGGGUAAUACAUAAAAACUCAAAACGACCC
GACGGAGCUGAGCAAGUAUAAAUUCUAACUUCCUAUACACAAAAGUAAAACUAUUUCAAG
AAUUGACUGAGACAUACACACACAACACAAACAAAUACAGGUCUCCGAUGAGCUAAGGGU
AAUAUAUAAAAACUCAGAAAAUGGCUCGAUGGAGCUGAACAAGUAUACAUCCU
2. MFE structure-
.............(((((((((................((((..(((......)))..)))).((((.......))))(((((((...........))))))).((((......)))).....((((((........)))))).((((..((((.(((.((.((((...(((....)))......))))))))).)))).
.))))................(((((((.(((((..(((............((((.((.....(((((....)))))...((((...(((......)))..))))..............(((((..........))))).....)).)))).))).))))).)))))))...((((((.............(((.(((((
........))))))))...............)))))).(((.((((...(((((.((.(((((..........)))))))...(((((.(((((((.........(((.........)))((((...............))))..)))))))...))))).(((....))).........((((((((.((.........
)).))))))))((((...(((....))))))).((((((.....)))))).........(((((.......(((((.(((((.(((((.(((....((((.(((((((((((..((((...(((((.((((.((.((((((..((((((((((((((((((............((((((((.........)))))))).(
((((.((.(((((((......)))))......)).)).))))).(((....))).......((((..(((((...((((((((.((((((((......(((...(((.(((((((........))))))).)))....)))...))))))))...)))))))))))))..)))).........((.(((((.((((..((
....(((...)))....))..)))))))))))(((((.(((((((..((.........))..))))))).)))))..........(((((........)))))..((((((.((((....)))))))))).(((((((((.((((((((....)))))))).))).....))))))................((((((.(
((...((((((...((((((.((((((((((.(((..........(((((((((((((((((((((((....((((....))))...)))))))...)))))))))))))..))).........)))))).))))))).))))))....))))))))))))))).))))))))))))))))))...)))))).)))))).
)))))))))...))))))))))).)))).....))).)))....)))))))))))))))))..))))).)))).)))......)))))))))......(((((((((..(((.((((......)))))))...(((((((((((((...((.((((..((((((((((......))))......................
.......(((((.((.....)).))))).......(((((.((.(((((((..(((((((((.........................)))))..))))..))).......((.(((((((..((....(((.....))).....((((((((..(((((((..(((((((.((((((..(((((.(((((...(((.(((
.((((.(((((((((.........((((...((((.(((......)))))))......)))).......))))))))).))))(((((((((((((((...........(((((....)))))((....((((....))))...)).((((((....((((((...)))))).))))))...(((....(((((((((((
(((.......))))))))....))))))....)))))))))............))))))))))))..))).)))))....))).))....)))))).)))))))..)))....(((.(((.....))).)))(((((((((((....))).)).))))))...(((((((......)))))))))))..))))))))...
...((((((((((.............((((((.................((((.......(((((((...(((((............)))))....)))))))....................)))).............................(((((((((((((.((..(((((....(((...((((((((((.
..(((...(((((.......((((...((.(((.........))).))...(((((.((((.........(((((.....)))))..((((...............((((...))))((....))...........................................................................
))))..........))))))))).......((((..................)))).(((((.....))))).)))))))))..)))(((((.....((((((((.((((((........))))))...))))))..)).....)))))............((((.....)))).))))))))))..)))))))).....
.......................((.(((((.......))))).)).......((((((..((((((......((((......))))(((.((((((....)))))).))).((((.........))))...........))))))...))))))....))))))))))))).))((((((((((.(((.........))
)..))))))))))))))))..))))))))))....))..))))))).))...........................))))))..)))))..((((.....................))))))))))..)))))))))))))))))))........(((......)))......(((((.........)))))........
.................(((..(((((.(((((((.((((...........))))......))))))).))))))))....))))...)))))
You can download the minimum free energy (MFE) structure in
II. Results for thermodynamic ensemble prediction
1. The free energy of the thermodynamic ensemble is [-706.92] kcal/mol.
2. The frequency of mfe structure in ensemble 1.84917e-42.
3. The ensemble diversity 723.11.
4. You may look at the dot plot containing the base pair probabilities [below]
III. Graphical output
1. A note on base-pairing probabilities:--
The structure below is colored by base-pairing probabilities. For unpaired regions the color denotes the probability of being unpaired. The reliability information of RNA secondary structure
plot in the form of color annotation, Positional entropy ranging from red (low entropy, well-defined) via green to blue and
violet (high entropy, ill-defined).
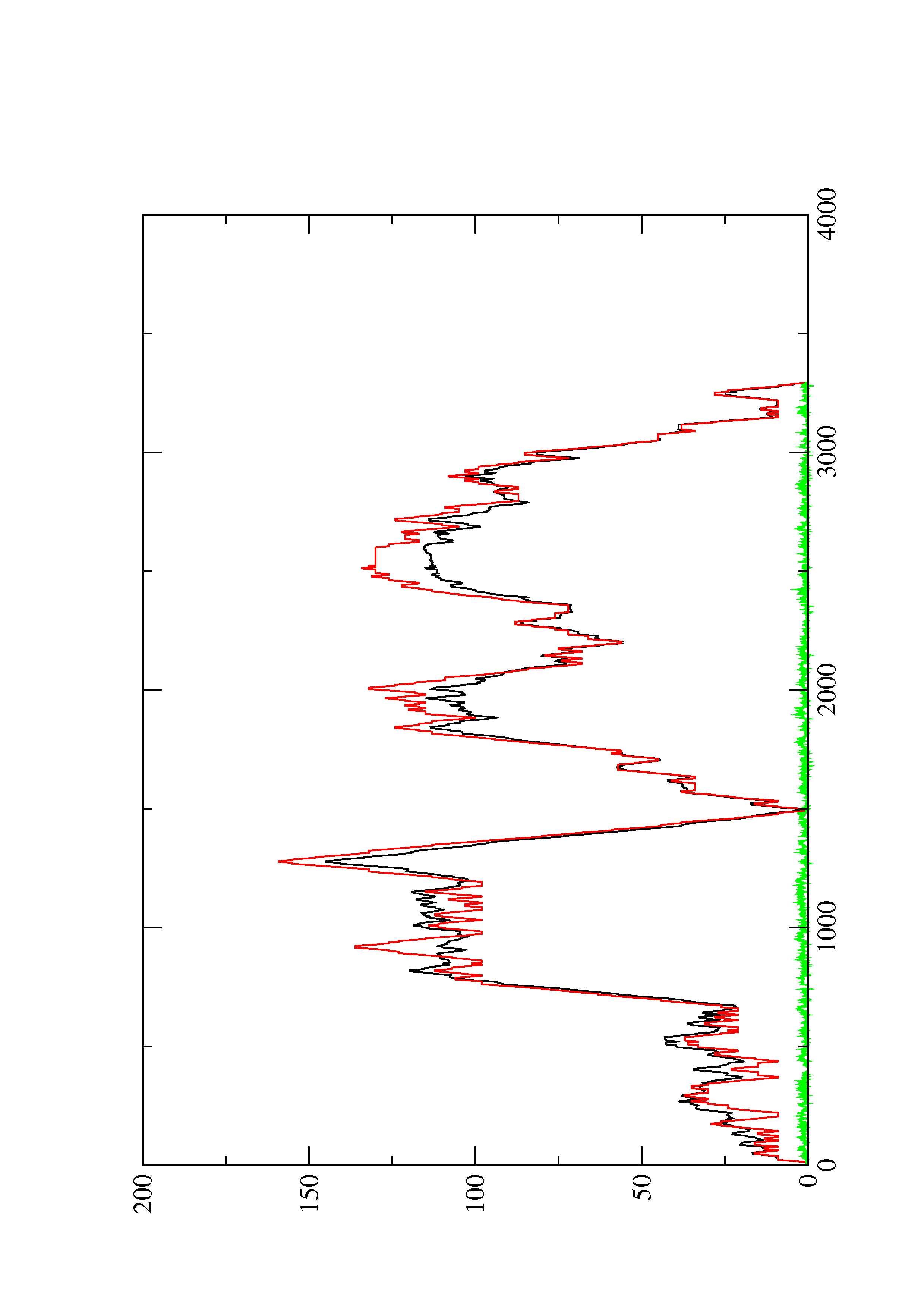
2. Here you find a mountain plot representation of the MFE structure, the thermodynamic ensemble of RNA structures, and the centroid structure. Additionally we present the positional entropy for each position. The resulting plot shows three curves, two mountain plots derived from the MFE structure (red) and the pairing probabilities (black) and a positional entropy curve (green). Well-defined regions are identified by low entropy.