lncRNA Structure Prediction
Species Name : Withania somnifera Sequence Id : GBHJ01018826.1
I. Result for minimum free energy prediction
The optimal secondary structure in dot-bracket notation with a minimum free energy of (-655.14) is given below.
1. Sequence-
AAUCAAUUAAGGGUCGCGUUGCGAGCUUUAUGCCAACCGAAGACUGGUGCAUCAUGGAUC
AAAGUACCAAUGCAAUCAAGUUCGAAAACAAUCCCAAACACUCCACGUUUAAUCACUACA
AGGCUGCCAUUAGCUCACAAAACAAAUCGAGCGCAGGAUUAUUGGGCAUCUCGUUUUGAG
UUUUUCUUGCCAAUUUGCUUCACUUUGAGCUUGAGUCUUGAUCCCAUCUCCUUCCUAACU
UGUACUAGGUUGCAAAUUGUUGCCAAAGCAAGCUCCGGAUUCUUGCAUUGCACAAGCCUU
UAUUUAUAGCAUCAAAGAUAGCCACACACAAGCUAACUAGCCUUAACUAGUUAUGCUUGU
GGAUUUUGCUCCUUCUUUCCUCUAUCUAUUUGCUUGUGUCAAUCAAAAUAUAGCUUGCAA
AUCAGUUCCAACAUCAUCCUACACAAGCUAAUAACAAACCAUCAGGCCAUUCAACACACA
AAAUUCACAUGGUACACACUAAAGCAAGCCUAGACUAUCCGAUUUAAACUAGUGAUCCCG
UCUAAAUUUUAUUUUCUCAUUCUUGCACUUGUUUUGGACCAAAUGUUCUAUAAACAUGAU
AACCAAGGUACUUAACACAUAUUUACACCAUUAUUCCCAUUUAAACUCGCAAACCCUCCA
AGUUAUCCUCAAAACAAAACUAAAAGGGUCGUUAACACAAGGUAAACAAGUGUCUACUUG
CACCUAUGAUCAACCACCCUUUACAUUCCCCAUCCUAUCACUAAUAGUCGACAUACUCCC
AUAUAAGUUUGACAUUUUUCCAUCAAAAGCACCAUCCAAUGAUUGAUAUUCAUAAUUGCC
UCCAUUAGAGAUCCCAUAGUAAUUUCUCCCAUGGGAGCUUGAUUUUGUCUCCCUAUGCCA
UGGUACCUAUUAAGAAAGACACUCAACAAAACAAGCAAAUACGUUAGGUGGUUAAAAGAU
AACCUCACAUGUCUCUCAAGUACACACCUUAUGAGUAGUCCUCACAAUUGGCGUGGCCAU
GUUUAGACUUGUUUUACUUGAUGUGCCACAAGGUGUUCAAUAUCUACUCGGAGUUCAAAG
UGAUUCUUGUUUUGAAGAUCAAUGGAAGACUCAAAAAAAGGGACUUGAGCCAAGAAUUCU
UUUUGAAACCGGUAACUGUACUUGAGCCAAAAAAUCAACAAUAAGACUACACAAGUAAAG
CACAACAGAAAUUGCACUUACCAGCAACAACCAAGUAAGAACAAUUGCUAGUUGAUAAUU
AGUUUAUGAAUCAACAAACGAAACUAGAAAGAUCGAGAAACUAAAAUUCCGGUGUCAAUA
UGGGUGAUCACUUGAAACAUGCUCUAAAUGAUCCCACUAACCUAUGGUGUCAAAAUAAAG
UGCUCCGUGAAGCAGACCCAACAAAAGAAAUGCCGGUCUGUCAAGAUGUGUCGUCAAAUU
UUAGCUACAGGCUCACUGUUGUGAGUGAUGGUUGGGAUGACGGUCCGUCACUGGUUCGAC
AGUCCGUUAGGUUGCUCUAUCAAAUUGAACUCAAGUUCAGUCCCACAGUUUAAGGACGAC
GCCAUGGUUGAUGGCUCAUCAUGGACUCGAUGUCCGUCACAUCUUUCUAUGCUCUGAGAA
UUUAGCUACAACUUUCGAGUUUUGGAUUUUCUUGGGCCCACUUCACCUAGUUUUUACCAA
UUUUUGGUCUAAAAACACCUUUUCUCAAACUUGGAUUAAUAUUUUUUAUCAAACUUCUAC
UUUUUGCUUCUUUUUCUUCUAGAACUUUAAAAAUCUUCAAACAACUUGAAAUUUUCAAAU
UUCCAACUGCUUCAUAUGAACUCAAAAAGACCCUAUAUUUCAAAUAUAGAUUCCUCUUAA
CAUGGAACAAACCCUAUAACUUUUCAAGAUCAAACAAUAACCAAAUUUCAAAACUCAAUU
CGAGUUCUUCAAGCUCUUUCAAGUCAACAAUGGUGAAUCACACAAACCAACCUGGAAUGA
CUCGAAAUUGAAGAUUGAGACUCAAAAUCACAAGGGAAUCAAAUUCUAGUGAUCAAAAUC
AACAAAUAAACACAAAAAAAUAACUUUUUUUUAUUUUUUUAUUUUCAAGAUAAGAAAACC
AAGAUUUGAAAUCAUAGGAACGAUCCAAUACGGCUCUAAUACUAUAUGAUGCAAAUCAAG
AUAUGGAUACGGAAGCGGAAAUACAACAAACGAAAGAUAAAAAUAGAUAAGGAGGGGAAA
AGACACAAACAAUGAAAUCUAUAGGCAACCAACAGUACAAGAGUAAAGAGCUCCUAUUAU
UAUGUUCAAAGAAGCACCAAUUCUAUCAAAUCAAGAUUAGACCCAACGAAUUAAGAACAA
UAAUCAUAUAUGUUCGACUAAGAACCUAAUCUGUAGGUGAAAGGAAUGGAUACCAAGCAA
CCACACCUUACAAAUCAACAUCAACAAGGCUUGGCAACCACUCAAUUCUCUCACCAGAGG
UCUCAUAAUUCAACAUAAUCUACUUAACAUCUAUCUAUAGUGCAAAAAACACUUAUUACA
AAAAUAGCCCUAAAGUAGCGUUGGAGGAAUUAAAAAAACUAGCUAUAAACUUGGACUCUG
CAACUUGACAUUGCACUUGAUGGGCCGUCGUGAUGUUGACGGUCCAUCUUCCACCGUCGU
CCAAACUAGCAAGCAGGCAACCUCAUCAAAUUUGACGCCAUGAGUGACGGGCCUUCGUGA
GUUCGACGGUCUGUCGUCCCUGCCGCCAGCCUCCUUUUUGCAUGCGACUAUCUGCACAUG
AUAUUGACCAGUAGAAGCUUGAAAAGAACCCUUCAAGGGCCGUCCUUGAUUCUCCACGUU
GGGUUUAGCUUCAAGAGUUGUGAUAGCGCCGGCCGGUAUUACAUCAAUAAGGAGGUUGUU
CAGUAAUCACCGAUAGUGGGAAGAAGAAAAUCCUUCUCUUGAAGGGGAAGGAAAAAGAAG
GUCCUAAUGU
2. MFE structure-
...((((((((((((.((..((..(((((((((..((((.....))))))))...((((((.((.((........(((((((((((.........((((.......))))...........(((.......((((((..((((.....((((.(((.....))).)).))..))))))))))......))).........
...))))))))))))))))))))).........((((........))))..((((....)))).)))))..))..)))))))))).)))).....((((((.......(((((...))).))....((((((((((((((......))))))).)))))))..........(((((((((.(((((((((((((((((((
(((((.....(((((((..........................)))))))...............(((((..................)))))..........(((((...(((..((..((((((.((..........)).))))))..))..)))....)))))..(((((((((.....(((((.....)))))(((
(((..(((..............................................)))....)))))).))))))))).....(((((((.(((......((((...(((((....))))).))))......))).)))))))........................(((((...(((........)))))))).......
...................))))))))..........((.(((((..((((.............))))..))))).))((((...(((((..((.....((((...))))..))..))))).))))....))))))))))).....((((((((...(((((((((.(((((...((((((...(((((..(((..((((
...(((..((((...)))))))...))))(((((((((((.((((((....)))(((((((((.(((.(((..(((((((.((((((((..((((...)))).......(((((..........))))).))))))))..))))))).))))))...))).)))))).....................))))))))))))
))((((((.(((((..(((((.............)))))..))))).)).)))).....((((((...............)))))).....((((.(((.......))))))).)))....)))))...)))))).))))).......((((.(((........))).)))).........((((..(((((((((((((
((..(((...(((.((((((((((((((.(((((((((.....(((...((((((((......))))).(((.((((((((((.((((((((....(((..((((((..((((((.((((.....(((((....)))))((((..........))))((.(((((.....)))))))...((((((.....))))))...
..........((((.((((((((((....((((....))))))))))))))..))))............(((((((((((...)))))..))))))..((((((((...((((..(((....((((((((..........(((((((((((...((((((.(((.(((.(((((.(((((.....)))))))))).))).
)))....(((.(((((((..............(((((((...))))))).((((.........)))).....(((((....(((((.((((.....................((((.....))))(((((((....((((.(((((.((.((((...........))))...))...))))).)))))))))))((((..
.)))).(((((...(((((...))))).)))))....................(((((((.....))))))).(((((((((((...)))))))))))....)))))))))..)))))...((.((.....)).))......))))))).))).....)))...)))...)))))))))))..............)))))
)))))).))))...))))))))..................))))))))))..........((((.....)))).........((((......))))(((.((((.(((.....(((((((..............((((((((....))).)))))..........))))))).....))).)))).)))...((((((..
............................))))))....((((((..(((((....))))).....((((((....((((((((((.....((((....((((.......))))..........)))).....)))))).))))...))))))......((.((((.....((((((..(((((.......)))))...((
((((((((((......))))))))))))..........))))))..)))).))..(((....(((.......))).))).))))))))))))..)))..)))..))))).)))))))))).)))))))))......))))...)))))))))).....)))))..)))).)))....)))....((((....((((((..
...))))))))))....))))))))).)))))).))))(((((((.((.....)).)))))))........)))))))))...))))))))))))).)))))))))....(((((((((....)))))))))....))))))........
You can download the minimum free energy (MFE) structure in
II. Results for thermodynamic ensemble prediction
1. The free energy of the thermodynamic ensemble is [-704.06] kcal/mol.
2. The frequency of mfe structure in ensemble 3.39351e-35.
3. The ensemble diversity 577.71.
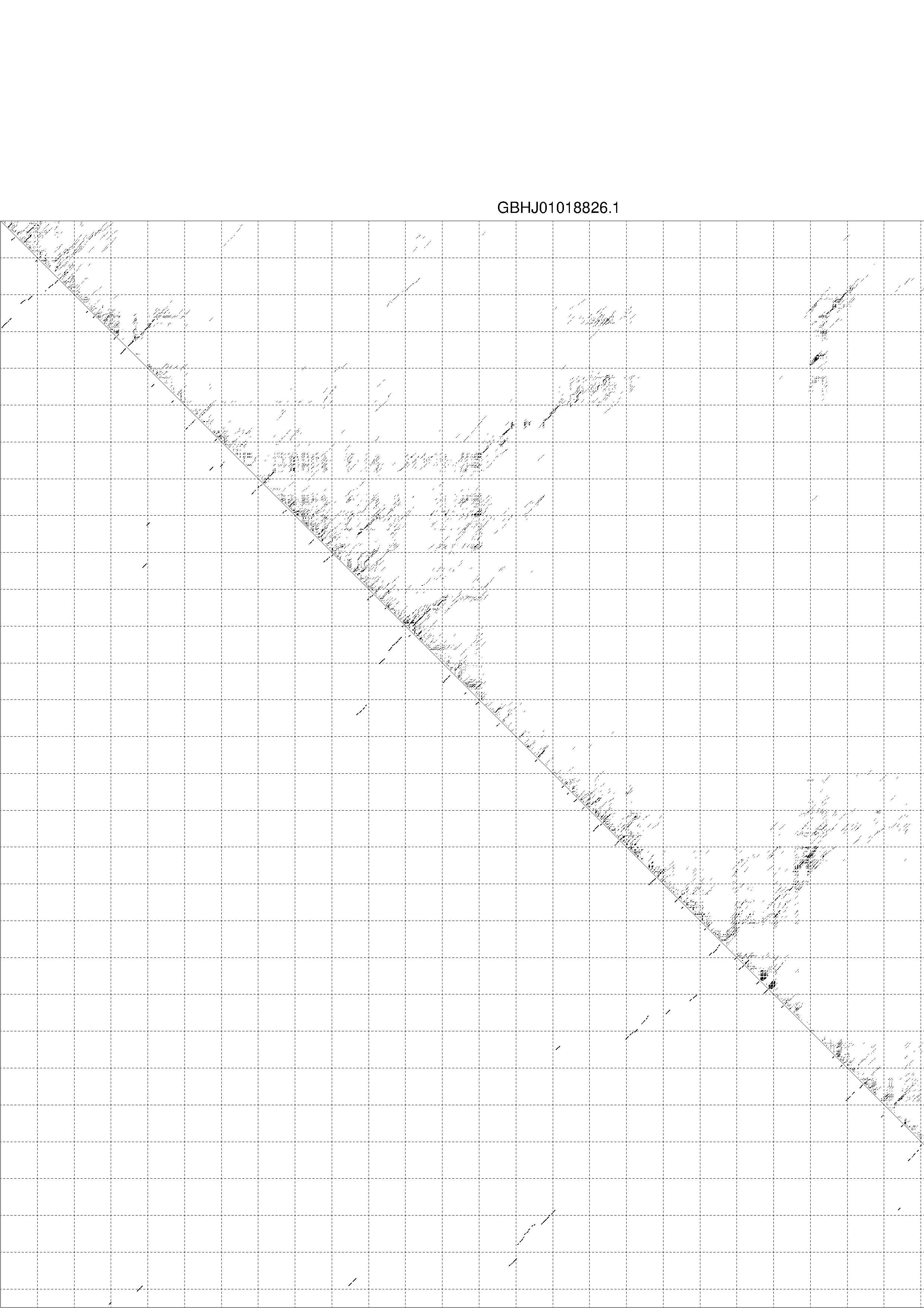
4. You may look at the dot plot containing the base pair probabilities [below]
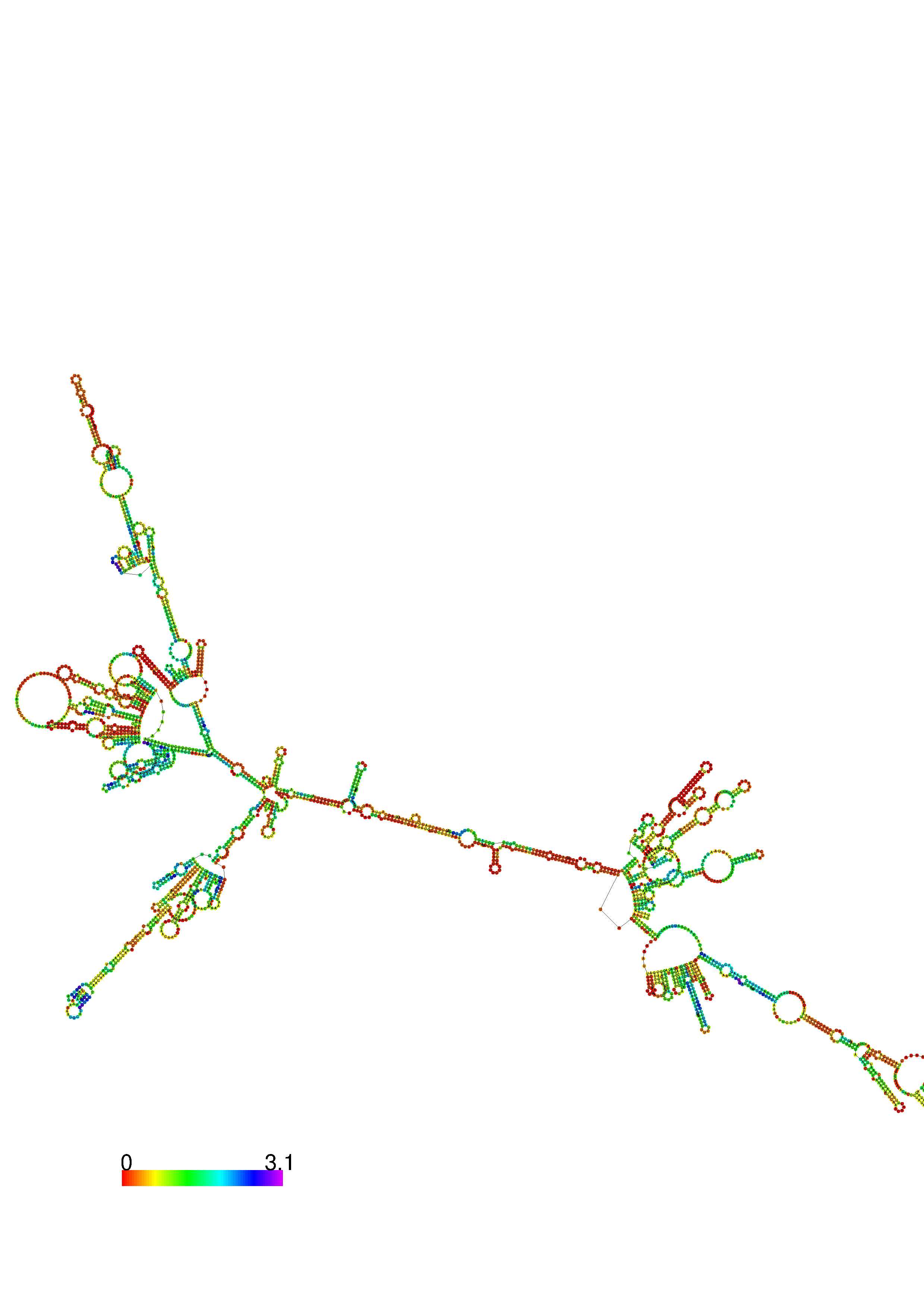
III. Graphical output
1. A note on base-pairing probabilities:--
The structure below is colored by base-pairing probabilities. For unpaired regions the color denotes the probability of being unpaired. The reliability information of RNA secondary structure
plot in the form of color annotation, Positional entropy ranging from red (low entropy, well-defined) via green to blue and
violet (high entropy, ill-defined).
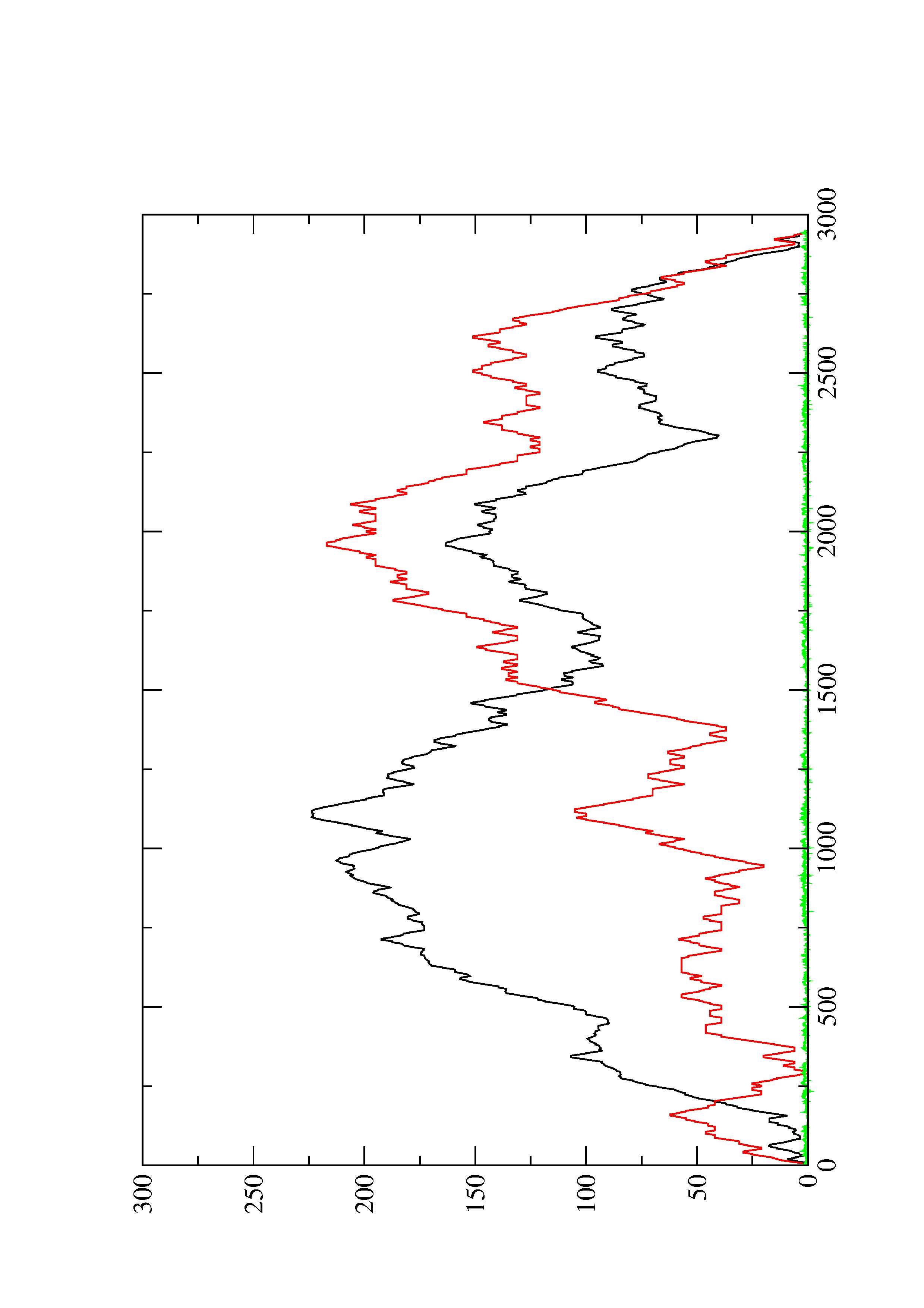
2. Here you find a mountain plot representation of the MFE structure, the thermodynamic ensemble of RNA structures, and the centroid structure. Additionally we present the positional entropy for each position. The resulting plot shows three curves, two mountain plots derived from the MFE structure (red) and the pairing probabilities (black) and a positional entropy curve (green). Well-defined regions are identified by low entropy.