lncRNA Structure Prediction
Species Name : Withania somnifera Sequence Id : GBHJ01019164.1
I. Result for minimum free energy prediction
The optimal secondary structure in dot-bracket notation with a minimum free energy of (-395.30) is given below.
1. Sequence-
AGCAAAUAAAUAAACAUAAAUUGUAAUCAAACAAUAAUGAGAGAGGGAGAGAGACAUCGA
UUGGGGAUUUUGGCCCAAACCGAAGCCACUUGGUCCAAUUCUCCUUAUCUAUACGCAUGA
GAGAGAACCGAAUCCACUACUUAUGGGUAUUUGACCCAUUCAUUUUCCACCUAUCCUAAU
UCUAACAAAAAUAAUAGACAAUAAAGAAAUUGGGUCUUAUGUCCAAUUACACACCACUAA
AUAAAGAAUAAAGAAAGGGAAUAAAUGACACCUAAAUGUGCUUCGCAACCCUUAUAACUA
UUGAUGAGAAAAAAUUGGAAUACUUUAGGCUUCUUCAAUUCUUCAACUCUUGAUCUUCAA
UUCAUUUAGGCUUGGCAUAGGAGGUUUGACUCAAAGUAAAUCAAAAGAGAGAGAAGAGCC
CACUUGGACUCGGAUGGGAGGAGUUCAUGCAAAAAAAAAAGAGGAGAAAUUAAUUAGAUA
UCUCAUGAAACUGACAACUAAAAGUACCAGCUAGUAAACAAACAUUUUGAUCUGAACAAC
CAAGUUGACAGCUUGAUUUUAGGGAUCCUGAACAACCUAAUUCCUAGUUAAACUCUCAUG
CUCCUAUUCAAAUAAAUGUAACAAUCAACAUGUUUCGCUAAUCCAAGUAGAAGGGAAACA
GAGAAAACUUUAACACAAAGAUUCAGCAAACAAGUAUCACCUCAAUGUUUACAAAGUCAA
CUACACACGGAAAGAGAGAAUUUGGAAGAAAAACCCAUAGACAAACCUUAAACAAGUUUG
AAACAAACAAACAUCAACCCACCUAUUUUUUCUAAGCAUGCAGGUUUCAUGGCACUACUC
AGCAAGUGACAAAAACUAUGCAACUAUAAUGCAUCAAAGGUUCUAAUUGAGCAAAUAUGC
AGGGAUUGACAGUAAACAGGGCUGGACGAGUUGCCAAAGGCUGAUUUUAACACCAAGACU
CUUUAUUUACUCAAAGGAAAUUCAAUUAGAACAAAACAGUGCUAAGACAUGUCAUUAUUA
CAUAUUGGUUAUAACACGCUUUGUAAAGUGUAAAAACAAUCAGUUUUGAAAUCAAAUAGA
CAAGUCAGGCUUGAUGGAAACAAAAGAAAUCAUAGGGACCAUUAGUAACUUGGAAAGUAG
GUUAGGAAUAACUUGAUCAUGGACAAACACCUCAAAUUUUAUCAAAGUUUAAACUAGCAC
AUGUUCAAAUAGGUGGCCGCAACCAAAGAGUUCUGCCAACAUUCUCGAAGACAAGCUCAA
CAGUUAAAGAUGAGAACUUUACUCAUUUUCCUCAUUUAGAAUGUAUACAAAUGGACAUCA
AAUUGAAUAGACCUCAUUGAACAGGUUGUUGCACAGGCUGUUGCACGCAUAUAUUAAACU
UAAAAGAUCCAAUUUUGUUUAACAACAUUCUUAAGUACCCAGACAGCGGAACACAUCAAA
CAAUACUUCAUUAAACCGAUAAAGAUCACAACUUUGACAUGGAAUUGAAAUAAUUCCUUU
AAGAGGGUAUCAAAUUUAGAUAACCUACAUAAGCACAUAUCCCGGUAACAAGUAAACCAU
UAGCAUACAUGUCUUAUGCAUCCAAUACAAAGCUGAUUGGGAACUAGUCCUACACCUAGA
CUACAUAUUCCAUUCAAGCAUAGAAAUCAAAGGGUUAAGGCAGCCUACUCUCAAUAAGUC
AUUAUACAUUGAUCUUAUCCUCAAAGAUGCCUCAGUACCAGUUAACCCAUGCAAAUUCGC
AUAUCCAAGCAGUAAACAAAGUUGAGCAGCCCAUAAUUAUAAAAAAAAUUAACUAUUUUG
AGAGACAAAGAGGCAGGAAUGGUGUGUUUUGAAAAAGCAAAUAGGGACUCAAAUUAUAGG
AGUACAAAAUGACCAUAUUUUCCCUUAUAACAUGCUGGUCACAUAUCUCUAAGCGUUCAA
CAAAUCUAGCCAACACCUUCGAUCUACGAUGACUCGUUAAGAAAGAAUAAAAAAGAGCAA
GAUGACCAGCCCGGAAUGCAAAGACGUGGACGAACAGUUCACAGGAAACAAGGCUCUAAC
AAAACAAGUAUCAACUAACUAAUCUUCUUAUGUGUACCAGAUUAUGUAUUUAGAAAAUUA
CGAAACAUGGAUACGCACAAUUAUUUACCCAAACGUGUUAAAAC
2. MFE structure-
............(((((..(((((......))))).....(((.(((((.((((....(((((((((((((((.((((...........)))).))))((((((((........(((((((.((........))........((((((.....)))))).....((((.(((((((....((((..........))))..
.........(((((((..((.(((.((..((.....((((((.((((...((((..((((((....((..((((((.(((.(((.(((..(((((........)))))......)))))))))))))))..))....))))))....))))..))))......))))))...))..)).))).))..)))))))......
...............(((.((....)).)))))))))).)))))))))))..........)))))))).....((((((.....(((....((...(((...(((....))))))...))..)))....)))))).....((((((...))))))..((((((...))))))......)))))))))))....))))...
))))).)))((((((.(((....(((..(((((((((.((((........((((((((....(((..((((.....)))).)))...............(((...(((((......((((..((((((..............(((.........)))..................(((((.......)))))......((
((((((((.......((((...((((.(((((((((........)))).......))))).))))...)))).......((((((((((((((....))).((..((((((((......(((..........)))....))))...))))..)).....(((((........)))))....)))))))))))..((((((
((((.(((.((((((((((.((.(((((((.....((((((....))))))......)))))))..)).)))..))).)))).)))(((((((((((((..........((..((..((.....))..))..)).....(((((((.....)))))))..((.......)).....))))))).))))))....))))))
.)))).))))))))))..............(((((((....((((((((.....(((((((.(((((((.((.((.(((((.((.............(((((((..(((((((((..........((((..........(((((.((((...)))).))))).........))))............))))..)))))..
....)))))))...((.((........)).)).........................))))))).)).)).)))))))..(((((((...)))))))......(((.((((.......)))).))).................(((.........)))....(((((.......)))))............(((((.(((
((..(((((.........)))))....))))).)).)))............(((((((((((..((.......((((((((........))).)))))......)).)))))..........))))))((((......))))..................)))))))......(((((........))))).....))))
)))).......)))))))...)))))).))))..)))))...))).((((.........))))...............))))))))...((((((((((((((((..(((..((((((.....((((.((.......(((..(((((((....))))..)))..))).........)).))))..........)))))).
.))).((((((.....)))))).(((........))).........(((........)))........)))))).)))))..))))).........))))))))))))))))....))).))))))........))))).....
You can download the minimum free energy (MFE) structure in
II. Results for thermodynamic ensemble prediction
1. The free energy of the thermodynamic ensemble is [-433.92] kcal/mol.
2. The frequency of mfe structure in ensemble 6.1471e-28.
3. The ensemble diversity 548.54.
4. You may look at the dot plot containing the base pair probabilities [below]

III. Graphical output
1. A note on base-pairing probabilities:--
The structure below is colored by base-pairing probabilities. For unpaired regions the color denotes the probability of being unpaired. The reliability information of RNA secondary structure
plot in the form of color annotation, Positional entropy ranging from red (low entropy, well-defined) via green to blue and
violet (high entropy, ill-defined).
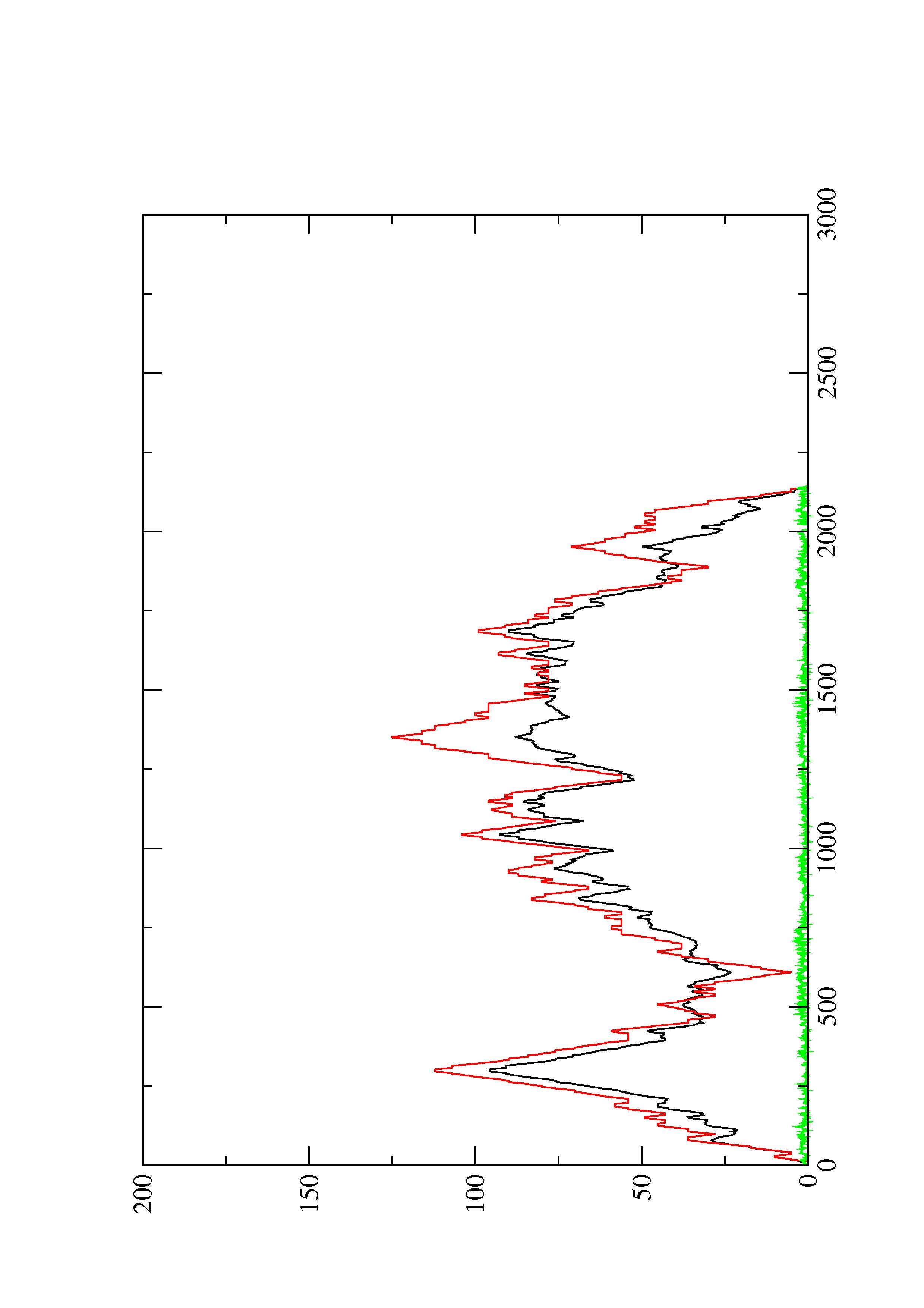
2. Here you find a mountain plot representation of the MFE structure, the thermodynamic ensemble of RNA structures, and the centroid structure. Additionally we present the positional entropy for each position. The resulting plot shows three curves, two mountain plots derived from the MFE structure (red) and the pairing probabilities (black) and a positional entropy curve (green). Well-defined regions are identified by low entropy.