lncRNA Structure Prediction
Species Name : Withania somnifera Sequence Id : GBHJ01019176.1
I. Result for minimum free energy prediction
The optimal secondary structure in dot-bracket notation with a minimum free energy of (-614.30) is given below.
1. Sequence-
AUUGAAUACUAUGGCUUUGUGAAGUGUUAUGAUUGAAAGUCUUGUUUUAUGUGAUGGUUU
CUCAUAAUUCUGUACUAGUCUAAAGGUUAUGUGUCUCAUUGUUAAAAUUCAUAUAUUGCA
AUGCAACUUGGUUACUUACUUGAUAAAUAUGAUUAUACUUGCUUAAGGUUUGAGUUUUUC
CUAUGUUUCUAUAUAUUGGAAGUAGGAUGUUCAAUUAGAAUGAUAAGUAAGGUUUCUCUC
UACUGCUGGCUUGCUAGUCCUUAUGGUUGCAUUGAACCUAUUUAAAUGGCCUGAUUGUUG
ACAAUAUUGAUCCCCGUCCCUCAGAUUAGAUGUAUGGUAUACUUACCUGAAAAGAUUAGA
GAAUUAAUGUUUGAAUACAUAAUUUGUGUAAUUUAUUUGCAUGUUUAUUUCUGCUUCUGC
UUCUUUUAAUCCUGUUGGUCACCAAUAGGUUCCUAACAGUAAACUAGUAGCUUGAAUUGU
CUGAUUACUGUCCUUAAUGAUUAUAUUUUGUUUGUUAUUGGCUUCUCCUAAGUACUAUUA
AUUCUAACCAGAACUCUUGUUAUUUAAAAUCUAGACUAAAGUCUUUAAGUUAUGCCUUGU
AUGAUUGACUUAGCAGUUAAAGUUAGUGAUUUAUCAGUUCAGCUUAUGAACCUGCUCCUU
AUUCAUUUUGUGUACUCCACUGUCCUGUUACACUUUGUGUUGGUCGUCUAGUUGUUUUUU
AACUCAGAUAUCAGUAUAUUAGACUAAAGCGCAUGAUUCAUUCUAUUAAUGUAUUAUCUU
GAAAAUAACUUAAGGUUGAGAUUUUUUUUUUUUUUUUUACUUCUACUUGUUCCUUGUAAG
AGCUGAGUUCAUUUCUUGUUAAAAGCACCAGAAUAAGUUUGGAAAUGAUUGCUUAAUGUG
UCUCAGUAGUAGUAGGAGGAUUAUUACCUAAUCUCUUCUUAAAUUAACUAAUAUGCAAAU
GUCCACCUACGAUUCACCUGAGCCAAUAUUGAAAAAAUGCACCUUUAGAAUUUAAAAUUA
UGUGUAAAAAAAGUUGUUUAUGAUAUAUUUGAGUUGAUUCCCUCCUACUGUGAUACAUAA
UCCAGCAUGCUCAUAUUUCAUUUUUUGUCAAGGUUAGUCUUCUGUUGGAAUCAAUAUUGC
UCCUGCUUCUCUGAAAACACAGUACUUGAGUUGUUAGUUACGUGAAGGUUGUGUUUAGUA
ACUUGCCUCCCACCUUUUUAACUUUGUAGGACUGCUUGAAAUAAUCUGACCUUAGAUAAU
ACUGUGUUUGCAUAACUGGCCUUUGUGUACUCAAGUGAAAGUGAUCUGAAUAUAGUCCUA
AAUAUCAUGUUGAUUCUAGUUUAUUUUCAAUUUGAUAUGUUAGUAUAUUUUCUGAGUGCC
CUCAUACUGUGACCUAAUUGAUGCGAAUCUAUAAAAUGUUGGGCAUGUUCUUUCAAAUGG
UAAUCCCUAAAUUACUUGUUAAUUGGUGUGGUGGAAUGACUUGAAUUCUGCAUGAUCAAG
UGAAUCUAGGAAACAUGCUAGCAUGUCAUAGACAUUAGAAUAUAAGGAUUCUCAGCCCAG
AAAUGCAUUACAGAAUGAACAUAUUCUUGUUGAUUACCGCUCCUGUUUGGUACAACAUGC
AUGCUUUAGCUUAUUUUGCAGUAGAAUCCAAGCUUGUUAUCCAUUCUAUGAAUUUUGGAU
GCAUAGUUGGUAGGGAUUUCUCCUUCUAAGAGUCUGCAUCCUACCCCAGUUUCAGCAUUU
GAAUGAUGAUUGGUUCAACUGGAAGCUUCCUGCCCAUGAAAUCUUUCCCUAACAAAUGAU
CCAUACAAAAGUUGGAGAUUCUGAAUCUGUUAAAGGAGUGUGAGACUCAUAAACCUGUUC
CUUAUGUGCUUUUCCUGCUCUGUGGAAUGACUUGCUAGCACGGAUGUCCGUUAGAGUUGA
AUCAAUUGAUGGAGCCCUGAAAUCUUCCCCAAAAUGCAUUAUGUGGUUCUAAUGAUUGCC
CUUAACAUGCCAGUCUAACAUGCGUUUAGAAUCACCCUUUUGUAAAGCCAAGUCGCACUA
UUUCAUGCUUUAUUUCCUUUCUCUGCUGAACAUGGAAUGUCUAUGGCUUAGAAAAUAUGA
AUCUGUAGCAUGGAGACUUUGUUUUCUUUGCUAGGAGGACCAGUACUCUCGUAUGGUUUA
AAGUCAAAAUCCUUCAUUUCACCAAUAUUUGUUUGAUCUGAAUCAUGACUGGGGUGAUGG
CCAUAGAUUAACAUAUUCUAUUAGUCGGCUAAUAGUAAAGUUAACAUAUUAAUCAUGCAC
UGUUAACUGGAUAAGACUUGUAUUAAUGUGUUUGAUUUAGUGUUGCAGUCAUUAAUGCUA
AUUGGUAUAGUAUGCCGCUCAUGCAUUUUUCCUAGUUAAGACAUAUGCGACGUGUUAAAA
UGGUGAGUCUAUAUCCUUAUUGCUAAAAAGAGGGUCUCAUGCCUGCAACUUUCUCUUACG
AGUAGUAACCCUGUGAUUCAAAUCUCGUAAUCAGUACCAUGAGCAUGCUCUCAGACUGAU
UUAGACUCACUAAAGGCUAUUGGAGAUUGUACACUUAUCUUAAAUAUGCUAACUGGAAGA
GGUUUGGCUGCAACCAGUUUGUUUGAUAAAGGCGUCUUCAAUCAUUUUUUUCUUCUUUAA
CAUAGUCAUUUUAGUAAAGUAUUCUCAAGCAUGUUCAAAUGUUGAAUGAAUAGAUACUAC
UCUGUUAAAUGUGUAACUCAUGUGCAGUUGUUUCUAUGAUUUAAUGAUUUAUCCAGGA
2. MFE structure-
...(((((((((((((.(((((((....(((((((((.(((((...(((((.((.....)).))))).(((((.(((((....((((((((.(((((((....((((((((((...((....(((((((((..........................((((((((..((((.((((..((((((..(((((.....))))
))))))).....)))).))))..)))))))).......((((((((.((((....))))....((((((.(((((...((((((...((((.(((....(((((...(((((.........((((((((((....((((....)))).....(((.((((((....((((.((((((((((...))))).)))))...))
)).....))))))..)))(((.((.......(((((((..(((....)))..)))))))......)).))).......)))))))..)))........)))))....)))))....))))))).....))))))...)))))..)))))).(((....)))...........((((....)))).(((((((.((..(((
(((.((((((.(((((....(((..((((((.....(((((.....)))))...........((((((((((.......(((..(((.....(((((((((....(((((........(((((((((((((...((((...((((...((((((((((.((((((....(((((((((((((.......)))))))....
................(((.((((((((((((..(((...(((.(((.....)))..)))....)))...))))))))..)))).)))....(((((((..(((((.(((.(((((.(((((((((..((((..........))))..))))).......)))).))))).)))...)))))...)))))))...)))))
)....))))))....))))))))))......))))...))))..))))))))))))).........(((.((((((((((.((((.((((..((((........(((((((((((((.....(((((....)))))..((....)).............((((.((((....(((((....(((.((((.((........
))..))))..)))....)))))....))))))))...........)))))))).)))))....))))..))))..))))...))))))).))))))((((((((.((((((.....................)))).)).))))))))....((((((((((((((.(((((((.((((((.(((..((((........(
((....)))))))..)))..))))))............((((((......))))))..(((.(((((...(((((((.......)))))))...))))).)))...))))))).))))))))))))))))))).....))))))))).....)))..)))........))))))))))(((......))).)))))).))
).))))).))).))))))))).))..)))))))....))))))))..))))).))))....))...))))))))))(((((((...((((.(((((....))))))))).....)))))))....((((((((((((((.....)))).))))...))))))..((.....))...........((((.(((((.((..(
(((.((....))))))........)).)))))..)))).))))))).))).)))))(((((......((.......)).....))))).....))))).)))))...........(((((((.(((((.((.((((..((..((((((..(((((((((...(((((..((((((((((......(((.((((((...((
(((...((((.(((...((((.(((.((((((.((..((((((((.((................)))))).))))..)).)).))))))).))))..))).))))..)))))((((((..((((((((....)))))))).))).)))....((((((((..((((((.......................)))))).))
)))))))))))).))).((((((((((.........))))).....)))))(((.((((...(((((((((((((.....((((.....)))).....)).)))))))))))...)))).))).))))))))))((((((.......)))))))))))..)))))))))..........(((((.......)))))....
(((((((((((.(((....(((......((((.((((((((.(((..........(((((((.....................))))))).)))...)))))).)).)))))))...))).)))))))))))..(((((((.(((((.........))))).)))).)))....))))))..)).)))).))..))))).
))))))).)))))...)))))))))......))))....))).))))))........)))))))............(((((((((((..(((((.((....((((...(((((.....)))))))))..)).))))).))))))).))))........
You can download the minimum free energy (MFE) structure in
II. Results for thermodynamic ensemble prediction
1. The free energy of the thermodynamic ensemble is [-665.45] kcal/mol.
2. The frequency of mfe structure in ensemble 9.01798e-37.
3. The ensemble diversity 760.38.
4. You may look at the dot plot containing the base pair probabilities [below]

III. Graphical output
1. A note on base-pairing probabilities:--
The structure below is colored by base-pairing probabilities. For unpaired regions the color denotes the probability of being unpaired. The reliability information of RNA secondary structure
plot in the form of color annotation, Positional entropy ranging from red (low entropy, well-defined) via green to blue and
violet (high entropy, ill-defined).
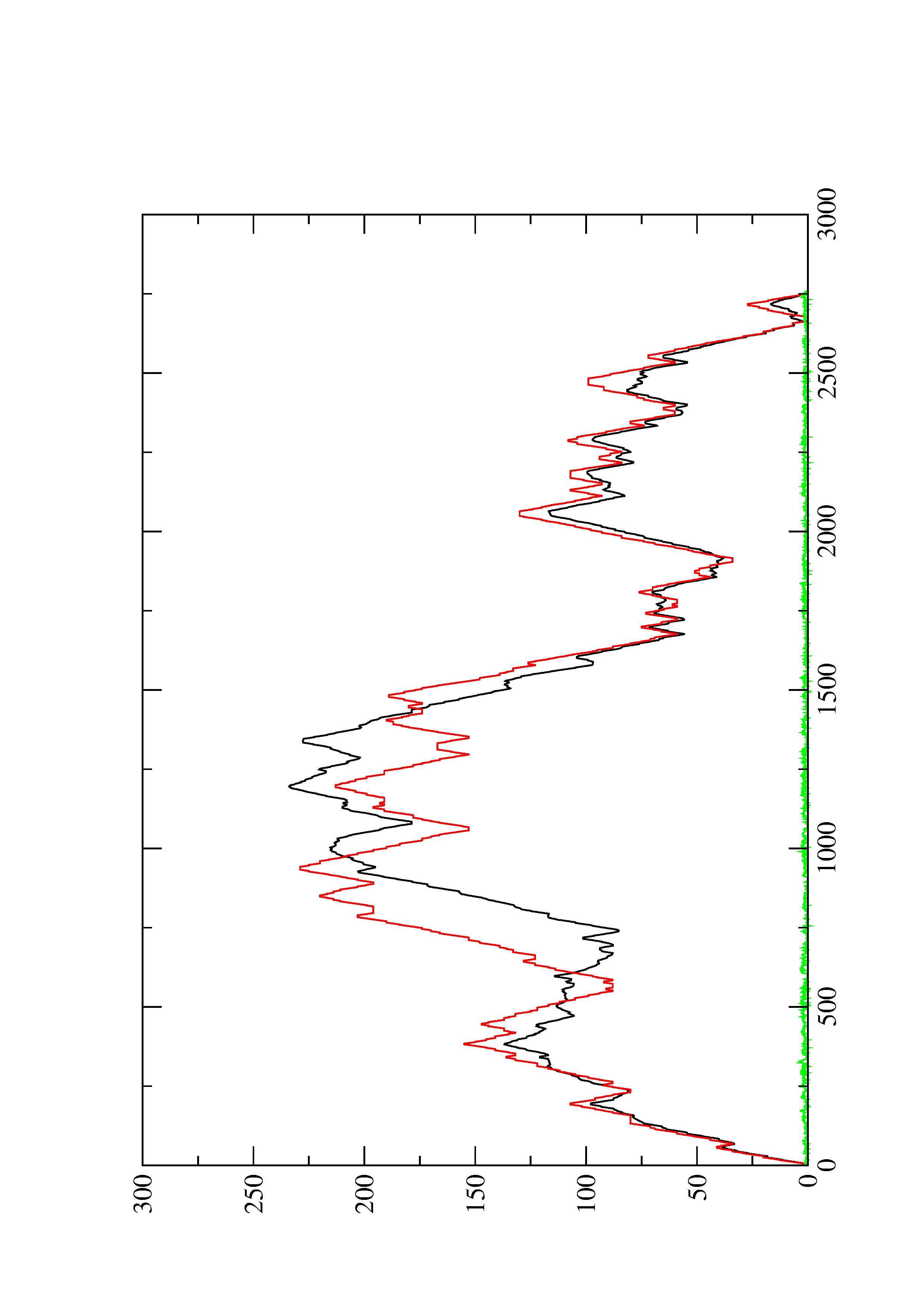
2. Here you find a mountain plot representation of the MFE structure, the thermodynamic ensemble of RNA structures, and the centroid structure. Additionally we present the positional entropy for each position. The resulting plot shows three curves, two mountain plots derived from the MFE structure (red) and the pairing probabilities (black) and a positional entropy curve (green). Well-defined regions are identified by low entropy.