lncRNA Structure Prediction
Species Name : Withania somnifera Sequence Id : GBHJ01019209.1
I. Result for minimum free energy prediction
The optimal secondary structure in dot-bracket notation with a minimum free energy of (-608.10) is given below.
1. Sequence-
GUUGUGGUUUGUUUUAUGAACAUCUGAGCUUGCAUUUAGGUUAGUAUAUGACUGGCAGGG
GUUUGUGUCUAGGAAAGUCUUAUUUUCUGUGUGAAACAUAUUAAUCUCCAGUAAAUGAUC
ACUUUGUGCUAUUUUAAAGUUGGUUCUCAUGAUGUUGAGGUUAAGUUUGACACCAUGCUU
GGGACAAAGGUAUUGUUUAACUUGAGUCCCUGUUUGCUUAGCCUGGGUCCCUAUUCAAGU
UUAAACCAUAUUGUUAAGUCCGCAUUCAUAAUGUGAAACAUGUUAUCCAUGUCUAUUGAU
UUGAAAUGUAUUCUCUUCUGUCUUGAACUUGAUCAUAAGGAUACUAGGGUAUCUUUUAGC
CUUGUUUACUGUGUGAAUGGGCAUGUUUAAGUCACCUGAUAACCUGAUCUUUUUUUUUCC
UCUAAUGCUAAACUGGAAUUUUUAAAGGAUUAUUUAACCGGAACAUUGUUUGCAUCUUAU
UCUAAGCCUAAGAGCAUAUUAGAGUUAUGUCAAAUGAGGUCUAGUUUGGUCUUUUAUUGA
UCUUGUAUCUCUGUCUAAAUGUAAGCUCAAUUGUGACUUGUUUAUGAAGUGUAUUCUGGU
UCCGAGGAGGUCUUUCUGUGAGUAAGAUGAUAGUCUUUGCCAUAGGAUCUCUACUGUUAC
CCAGAUCAUAUCAAAGUUGCAGUUUACCUGACUUAGUCUCCCAGUCAAGCCUAUUUGUUU
GAACUUCAUGGUAUUUUAAGUGUGUGGAUCUAAGCCCACGUCCUUUGUGUAAUGUGAUUA
AAGGCAGAGUCUAUUCAUUUGUAUGUUGUUUGCAUGCAUUUAGAAUGAGUAAACAGUCUU
ACAAACUUUGAUCAGUUGUAUAUCCCUAAUAUGAGCAGAUAAGAGUUUUAUGGUAGUUGG
UUUGAUAAAAAAAAAUAGGCUCUAUGGUAGUUGGUUAAUUGUGAAUUUUCUUUUGUUUCG
AAGAUGCCUAUUAGUCUUGGAUAACUUGCUGUCAAACUAAUGUCUAAGAUAUGUAGGUCG
UAGUUUCUGGUUUCUUAAAUCAUUGUUUCACUUGUCGAUAUAACAAAGAUAAGCUUUGUA
AGAUGAUUGGUCAUAAGCCUCGCUUCCUAAUUAAUUUUGUUGGCCUUAAUAAUCUAAUAA
AAAAACUGUCUUUACAAUGGAACUUAGGACUGGAAUGCUUGAACUCAUCUGUUUGGUGUC
AAAAUUGGUGUUUUUAUGGUCUAGCCAGUACCGGUAUCCUGUGUUUGCCUAUUUACUUAA
AUUGUAUGUUCCCUACAUGUUAGUUGGUGGAUAGACCCGUUUCAAAGCCUUGAACAUCUG
UUUAUCAGCGUGACUAGUUAAAUAUUCUGUGUUCGUUAAAUGGGAUCAGAACUGGCUUAU
AAAGUAUGGUUUAAGUGUUCAGUUCUCUGUUUCUUGGAUAAGUAAGGGCGAAUAGAAGUU
UUCUUGUGACACUUAGUAGUCUGAGGUAAGGCAGAGGAAGAUCCUUGAAUGAUAUGGGUC
CAAAUGAUCAUUCAGUAGUAAUUUUCUGUUCGUUUCCUUUACUCACUAUCUGGAGCAUGC
UAUAGAAUUAUAUAGUCCUUCGCCUUUUUGUCUGAUUGGGUCGAACAUUGAUGUCAUGGU
GAUCCUGUAUGCCUUUGUUUGUUUAUCAGAUAAGAAAAAACUGUUAUUUAGGAGUGAGGU
UAGAUCAUUUAGUGUUAAGAUCAAUCUCUAAUUGCUUGGUUGAGGCCUGCUUGUCUGUAU
AUUCUUCAAGUGACAGUAUCCGUGCUGGGUAAUAUGUUUGAUCAUGAUUUGAGUGACUUC
UGUUGGUUGCUACCAUUUUAUGUCAUGCUUAAGGCUUAAACAACACCUGAAUCCUCGUAU
CUUCAUCUUUUCUUUUACUGCGUCUUCAGCUCCUAGGAUGAGUUUACUAUCCUCAUAACC
CGUUGAAUUGAAAUUUUAUGUGAUUGUCUUUCAUCAUAAGCUGGUAUCUAAUCUCUCUCU
CUUUUAUUCUUGCGUGUGAUACCCCGCCUGAGUUCACUGUGGACUUUCUUCUUCUUGCAU
UUCUCUUGGGCCAAGGCCAUUUAUUCCCUGAGCUAAGCCCCAUCCAAGUCCUAUAUGGAC
UGAAGACAACAAGAAGACAAAGAGUAGAAGUGGCACGAAGAGCCGAAAAAUAAAAGCAGA
AGCAAUGGAGGCGGGCUAAAGGCCCAAUUUUGAUUUGGGACAGAUUAUGGAAAUUUGUGA
AAAGGUCAAACGCUCAUCAUUAGCAGCAGUAAUAAUUAGGCUUGAGAGCCCAAAACAUGG
UACUUAGAACAGUUGGAGGGUUGGGCCGAAGGCCUUGUCCAAUUUUCUUUCCCUCUACCU
AUAUUUGUUCGUUAUUGUUUGUUAUCUGCUGCGAACUUUAAACAUUUUAUCUAAACUUGG
UUGAUUCCCCAAAAGACAUACCUUCC
2. MFE structure-
.....(((.((((((.((.(((..((((((((((((((((..........(((((.(((....(((((...(((((((...))))))).......)))))....))))))))....((((((((((........))))).)))))...((((((((..((((((((((.(((..(((..((((((...(((((.((((((
((((((((((.(((.....)))..))).....))))))).)))))).))))).....)))).))..)))..))).))))..((((((((((((((((.((.......(((((((.....(((........)))....)))))))((((((........))))))........)).))))))))))............)))
))).(((((((((...((((..............)))).....))))))))).))))))..)))))))).(((.(((((.........))))))))...(((((.((((((((..((((.(((...))).))))..)))))...))).)))))))))))))))))))))..)))((((.(((((((((((((.....(((
((((((((((((((...((((((((((((.((((....(((((((((........(((((.(((((((((((((....(((..((((((((((....((.(((....(((((((((..((((((((((((.(((...(((..(((((........)))))..)))...))).)))))...(((((...((.(((((((((
((((((.....))))))).....)))))))).)).)))))..(((((((.(((.(((.(((((.....)))))))).))).)))))))..........))))))).......)))))))))...))).))..))))).......((((((.........))))))((((((..((((((((((.................
))))))))))..)))))).........((((((...)))))).....(((((((..((.(((....((((((((......(((.((((((((.(((((((((((.(((((.((((((((((((..........)))))))).............(((.(((((..(((((((.((((((((((((((((.((((......
.)))).)))).(((((((....(((((((...((((.........))))................((((.....))))(((((((((((((((((...(((.(((....)))))).))))))))))))).))))...........((((.((((........))))))))))))))))))))))...)))))))))))))
))))))..))))))))(((((((((((((((((((((((((((((.((((.(((((....)))))))....)).))))))))..(((((((((((........)).)))))))))........)))))))))...))))))))...))))(((((...((((((......)))))).)))))..((((((((((((....
.((((((...(((.((.((....))))))).....))))))...))))))))))))......)))).)))))))))))))..............))).)))))))))))....((((.....))))..))))))))...)))))..)))))))(((((....))))))))))..)))))))).)))))))).))))))))
)).)))).)))).)))))))).))))....))))))...............))))))..((((((((((((((((((........((((((.....((((((...((.(((..(((((...(((.......))).))))).))).))..))))))...))))))................((((((((((((((......
....)))..(((((((...)))))))...((((((((...((.((((((((...(((((.........)).)))..))))......(((((.....))))).)))).))))))))))..)))))))))))((((.......)))).....)))))).)))))..))))))).(((((...)))))...((((((((...(
((((((.....)))))))....))))))))............(((((((((((((.((((((....))))((((....((((...((((.(((((((.....(((((...)))))..))))))))))).......))))....))))........)).)))))).))))))))))))....))))))))..)))))..))
)).......)).)))))).)))....
You can download the minimum free energy (MFE) structure in
II. Results for thermodynamic ensemble prediction
1. The free energy of the thermodynamic ensemble is [-654.53] kcal/mol.
2. The frequency of mfe structure in ensemble 1.90257e-33.
3. The ensemble diversity 664.91.
4. You may look at the dot plot containing the base pair probabilities [below]

III. Graphical output
1. A note on base-pairing probabilities:--
The structure below is colored by base-pairing probabilities. For unpaired regions the color denotes the probability of being unpaired. The reliability information of RNA secondary structure
plot in the form of color annotation, Positional entropy ranging from red (low entropy, well-defined) via green to blue and
violet (high entropy, ill-defined).
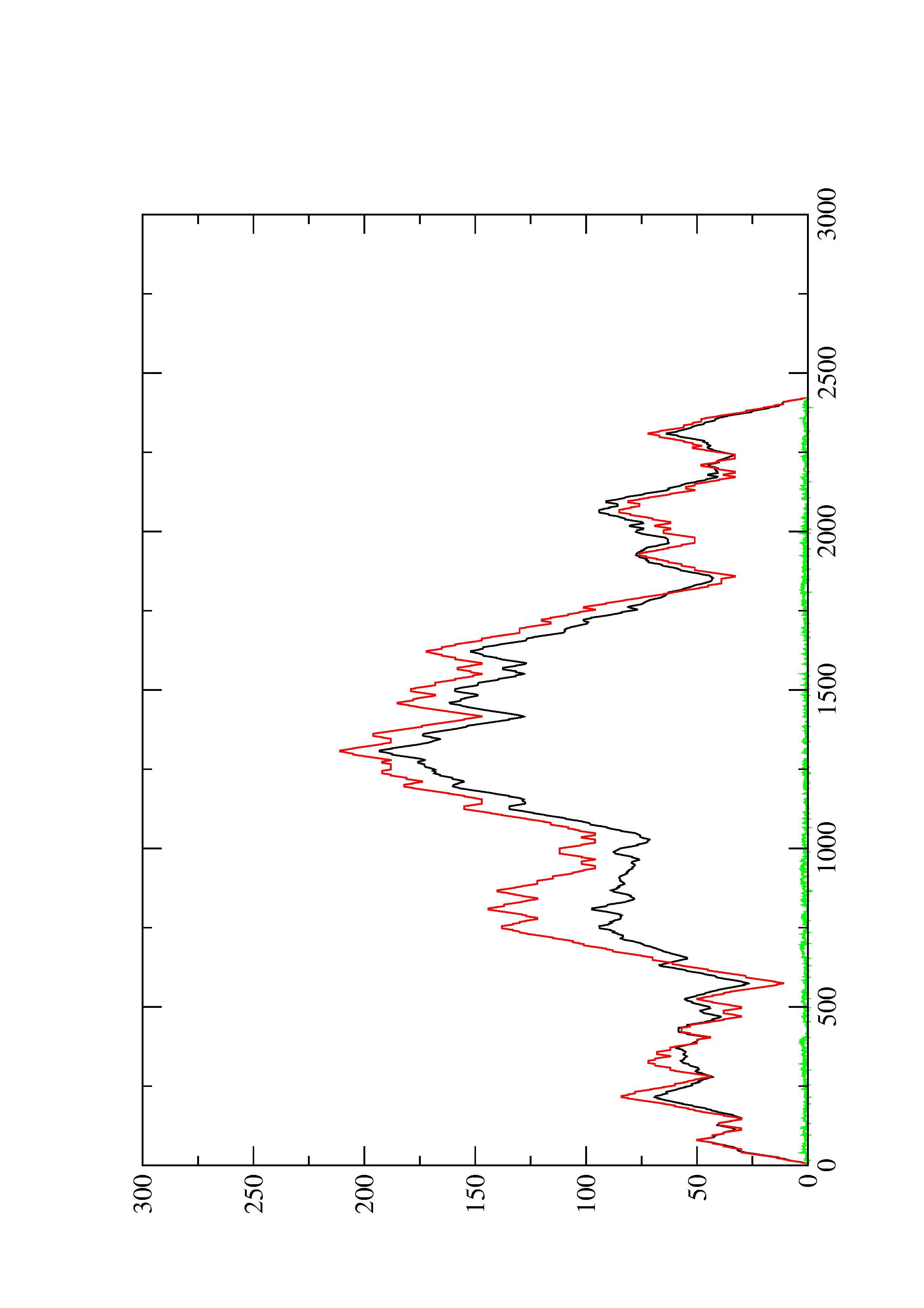
2. Here you find a mountain plot representation of the MFE structure, the thermodynamic ensemble of RNA structures, and the centroid structure. Additionally we present the positional entropy for each position. The resulting plot shows three curves, two mountain plots derived from the MFE structure (red) and the pairing probabilities (black) and a positional entropy curve (green). Well-defined regions are identified by low entropy.