lncRNA Structure Prediction
Species Name : Withania somnifera Sequence Id : GBHJ01019634.1
I. Result for minimum free energy prediction
The optimal secondary structure in dot-bracket notation with a minimum free energy of (-596.94) is given below.
1. Sequence-
AUUUGCAACUCAUGGGCCCCAUAUUUGAUUCUAUACAUCAUACAAUGAAUACAUAAAAUA
AAUAAAUGCAGAAAUGGUUUGGGUUGAGUCUCAAAGCCUCCAAUAGGGGCCAUAUCCCAU
AACACAAUGCGAAGAAGCCCAAAUUCACACGUGUAAAGAAAAAAGAAAUUAGAAAUCAAU
AAUAAUAGUUAUUACAAGUAAAAUAUAGUCAAUCGAGAAAGAAUAAUUUGAAGCUUAUGU
UGGGCUGGGCCCAAGAGCACACAGAGUGAUCCAAUAGAACAAGUCAGAAGGUGGCCCAAA
AUAAAUUCCAAUCGAUUAAAGGAAAAUGGGCUGAAGCAGAUUACAUACAGGUCAAAUCAA
UUUUAAUAAAGAUAUGUCUACCGUUUUGCAAAAUUGCAAUCCGACCCAACCUCUAAAUAA
UUCAACCUAAAUGAACAAAUAUCGCCCUAUUUAGCCCUAUAUCAAGAAAACAUGACUAGA
UAAUUAAAUUUAAUCUUAAUUACCUAAGUGAUACACAUACUAUACAUACAAGGUUAUUCA
UGAACAUUGAAAACAGAAACAAGUACAGAUAAUCAUUUUAAUCCGAAGACUAGCAGCAUA
AGAGUAUAUAAGAAGAAUGGGAGUAGAAGAGACAAGUAAAUAAGCAUCACAUAGAUACAU
CACAUACACUAUGUUGGGCCUCACUUAACAAACUUUCUUGAAAAUUAUAUCAAACAUAUG
UAUGUGACAUCAAAUAGGGAGGGCCAUGUCAAUGAUUCAAUAAAAAAAAACUGAGGAAGA
AGUAAGCAUAAUUAACAAAUAGAAGAAGAGAAAAUAACAGGUUACAUAUAGACAAUCACA
AUUCAAUAGUAUAUGUGACGAAGGGGAUAACUAGUCAGCUAUGGACUCACCAUACAAUGG
AAAAGAAAGACAGCAGCUUGAUGUUAUUAUAGACUCCAUACAAAGGGACAAGAGGUGAAU
AGCAUGACUGCAGGAGGUCAAUAUGAAUAGUAAAGAAAAACAGGUAGCAAUUUAUAUCAG
AGAAGGGAAAGCAUAUAACAAGCUUAUAAAAGGCACCAUAACUUAGCAUAGAAAAGGAAG
AGGCAUGUGUAGAUAGUUUAAACAUAACCGAGGAUAUACUGGACGGCAGGAUUGAGAGAA
AAUAGGUCAUUCAUUUAGAUGCACAUUACAAGUCUAAGGCAGCAACACAGAAGCAUGUAG
ACAUAAAAGAGUAACAGGUCAUCUAGUGCAAGUUUAACACAUAGCAGGCAAAGGAGUUUU
AAAGAUAGGCCAGUCAGGUUGCAAGUGAAGUUAUGUGCUUGUGUGCCAUAUGUGAACCUA
AGGCAACAACACAGGAGAAAAUACAUGCAUAGAAUGAACAGGUUAUACAGAAUAGGUCCA
UGGAAGAUAUAUAAUCACAUUGCGGCAGUCAUCAGAAGGACAAUAUAUGAAAAUAGUCCU
AUAAAAAAUAUCAUAUACCUAUAAGAGGGAAUUGAGACCUAGAAUAAGGCAGGCCUUAGA
CCGGCUACUAUAUUUGAACAAAUUUGACAUGAGGAGGCAGCUAAGAGUAGAACAAACAAC
GUAAAAAUCAAGAGAUCAUGUAUAUGGGGCAAUCAGACAGAUUGAUAUCAUGUAAUGUGA
AUUUAAUAUAGAAGAGGAAAGUGAGAAGGUUGCCAUAUGUAAUGGCAUAAAUCAAAAUAA
AGUCCAGAUAAGGAUGCAACAGAAAAAUCUAUAAUUAAAAUGAAUAAGGGCAGCAGACAA
CUAGUGUAUCUGGUAGUGCAUGAGCUAAACAUUGAUGGGAGGAUGAGGCUGGCUCAAGCA
CUGUCACAUAAGUUUAGCAUAAAGAGAAAGAAACAGGUAGCAUACUCACAAGUAAGCAUC
AUGAGUUUAAUAUAGAGGAAGAAAGAAAUCAGAUGAUCAGUUUGGCAAAGACUUACUUUA
AUUUUAAGCUACAGUACACAGUUAUUUAACUGAUGGGGGUUGAGGGAAGAAGAGCAUGCU
UAGUGGUAUAUAAUUUUUAAUAUGAGAUAACAGGUAGGCAAAGUAGUCAAAACAAGGGAC
UAUGAGUUAAAUAUUUAUAAUCCCAUGCACUAAACAAGAGAAAAUAUAAGAAAAGUGAUA
GGUUAGGCAGUUACAUAUACAACAGAACAAAACAAGUAGUCAUCAUGAUAGUUUCUAGAG
AAGAAAGUUUAUAAAGCAUGCUAGACCAAGACUUAAGGCAAAAAAUACCUACCCUAGGCA
UACUAUAUAUUAAACUUAGAAGGAAAACAUUUAAUCCUUAACCAAACAUAUAUAAAUAUG
CAAGUAAAGCUUCAAUAAAGCAGCAGGCAGAUUAUAUGGGAUCAAAUAAGGUCUUGUUAA
GCAGUAAACUGAGCAAUAUGAAAGAGUACAUGUAUAGACAACCCAAAAUACUAAAUGCUU
CACUCAACACAAAGCAAGUAGACAUUGUAAGUACAGUUACAGCCUAGUAUGUUAUGCACA
CAGUGAUUAGACUCAUUUAGUCCAGGUUUAGGCCAUUGCAUCAAUUUUAAGCACUAUGUU
AGUCAUUAUACAUAUAAGAGAAUGAACAUAUAACAGGUUAGAGGGUCAACAUGGACAUGU
AUGAAAGUGACAUAUAAACCUUACUCAUAUUAAGCAUGCAUGACAUCUCAGGGCUUCCAU
ACAACAUAUAUUUAGCAUCUAACAAGCAAGAAAGUUUAACCAGUCCCAAAAGGAUCGCCA
AAUACUUAACUUUCAAUUUCAAAAUGACACUAAGCAAGAAUCAGUUAGAAGCACUGAUUU
GAACAAACUACUUAGCCAUUCAUGGAGGACAAAGAAACAUACUACAACAUUAUUAGAGAA
AUAGUCUAGAAUAUGCAAAUCUAAGAGUGUAGCACGCAAGAUCAAAAACUCUAAAAUAAG
GAAAUUUUUGCUCUAAAAUCUUUAAAAAAAAAGCAUAGAGGAUAUUCCCACCCUAGAGAU
GGUCACACUCUUAGACUAAUUGGAGGUGGGUUGAACCCAUAUCAGCUCAUACCCCUCACG
UUAGGCAUCCUUACAAGAUUUAGGUUCAUAGGAUUGGUAAACAAGCAUUAAAUUUCCCAA
GUUUCAUCUCACAAGUCAUCUCCUUGCAUUGUACUCUCAUCAGUCAUUAUGCAUUAUAGG
UAAAAGAGGUGUGAAGUGAAUAAAUACUGAAUCUUCACUUCCUUUAAAAUCCUUUUGAAA
AUCAAAAAUACUCUAAGAGUACACUUAGAUAAAAAAAAUAUCAC
2. MFE structure-
............((((((((...(((((((((((..(((((...(((....)))............((((....(((((((((((...((.((..(((((.....))))).................)).))...)))))))).)))....))))......................((((((....)))))).(((((.
........((....)).........)))))..((((...((((((...)))))))))).......)))))...)))))....)))))).)).)))))).................(((((((((..(((((((...)))..........((.((.(((............))).)).))..((..(((((....))))).
.)).))))((((((..........(((((..((((((.((((.(((((......((((((.(((........))).(((.(((((((........))))))).))).(((((.....((((..........(((((((..((.((.(((..........))))).))))))))).(((((.(((................
...................))).)))))......)))).......)))))...(((...(((((((((.((((((.............(((......)))............))))))))))))))).)))..))))))))))).))))...)).))))..........(((........)))..(((((((........
.((.((.((((..((((...((((((((((...................)))))))))).((((((((.(((.((.((..(((((((..((((...))))............((((((((((....(((((((((((................((((.......(((((......)))))((((((((.((.........
......)).))))))))...........((((.........))))........))))........................((.((((.((((...)))).))))..)).........(((.....)))((.(((...........))).))..(((((((.((.((((..((((...(((((.........)).)))))
))........))))...)))))))))(((..(((........)))..)))..))))))).....))))....))))))))))....((((((..((((((((.(((((..((((((((((((..........((((.........((((((.((((....(((.............((((((((.(((.....)))....
(((..((((((((((((((((.((........)).)))))..........(((....(((.....)))....)))..(((......)))..((((.......))))......))))).......)))).)).....)))(((((((((...........................((((((((((((..(((((.....)
))))...)))))).)))))).(((((((((((.........(((......((((((.....))))))....))).......((((......))))............))))).))))))..........(((..((....))..)))...(((((((((.(((((((..((((........))))....))))))).)))
)))))).....(((((((((...((((..((((.(((((((.((((....)))).))....(((((((((((((.((............(((((.....)))))((.....((..((((((((((...........(((((((....))))).))))))))))))..))....))((((((((.(((.....(((((...
...)))))...(((((......((((((....((.((((.(((((((.....))))))).)))).))((((......................))))...((((.(((((((..(((.((..................((((...))))..(((((.....))))))).)))..))).)))).))))..))))....)).
.....)))))))))))))))).)).))))))))))))).(((((..........))))).......(((((....)))))........((((.....))))....(((.((((...((((((....((((.(((((((....((....(((((..(((((.....(((..(((((..((((.......((((...(((((
...........)))))))))....))))..)))))..)))(((((((.((((((((((...(((((......)))))..(((((.(((..((((..(((...........))).((((((..((.((((....)))))).....)))))).............))))))).)))))..((((((..((((..........
))))))))))......))))))))))))..))))).........))))).)))))...))....)))))))...))))....))))))....)))))))...)))))...))))..))))...)))...))))))..((((((((.......))))))))......))).))))))..))))))))..............
)))....))))..((((.......))))...))))))..(((((((((((.........(((((....(((((......((((.......(((((....((((......))))..)))))....)))).....))))).))))))))))))))))(((((..((((((((((((.......))))))))....))))..)
))))...)))).......))))))))))))....))))).))))))))..))))))...)).))))))).)).))).))))))))...))))..)))).))..)))))))))....)))))...))))))((((((................))))))))))))))).....((((((...))))))....((((((...
...))))))...............
You can download the minimum free energy (MFE) structure in
II. Results for thermodynamic ensemble prediction
1. The free energy of the thermodynamic ensemble is [-656.07] kcal/mol.
2. The frequency of mfe structure in ensemble 2.16884e-42.
3. The ensemble diversity 818.75.
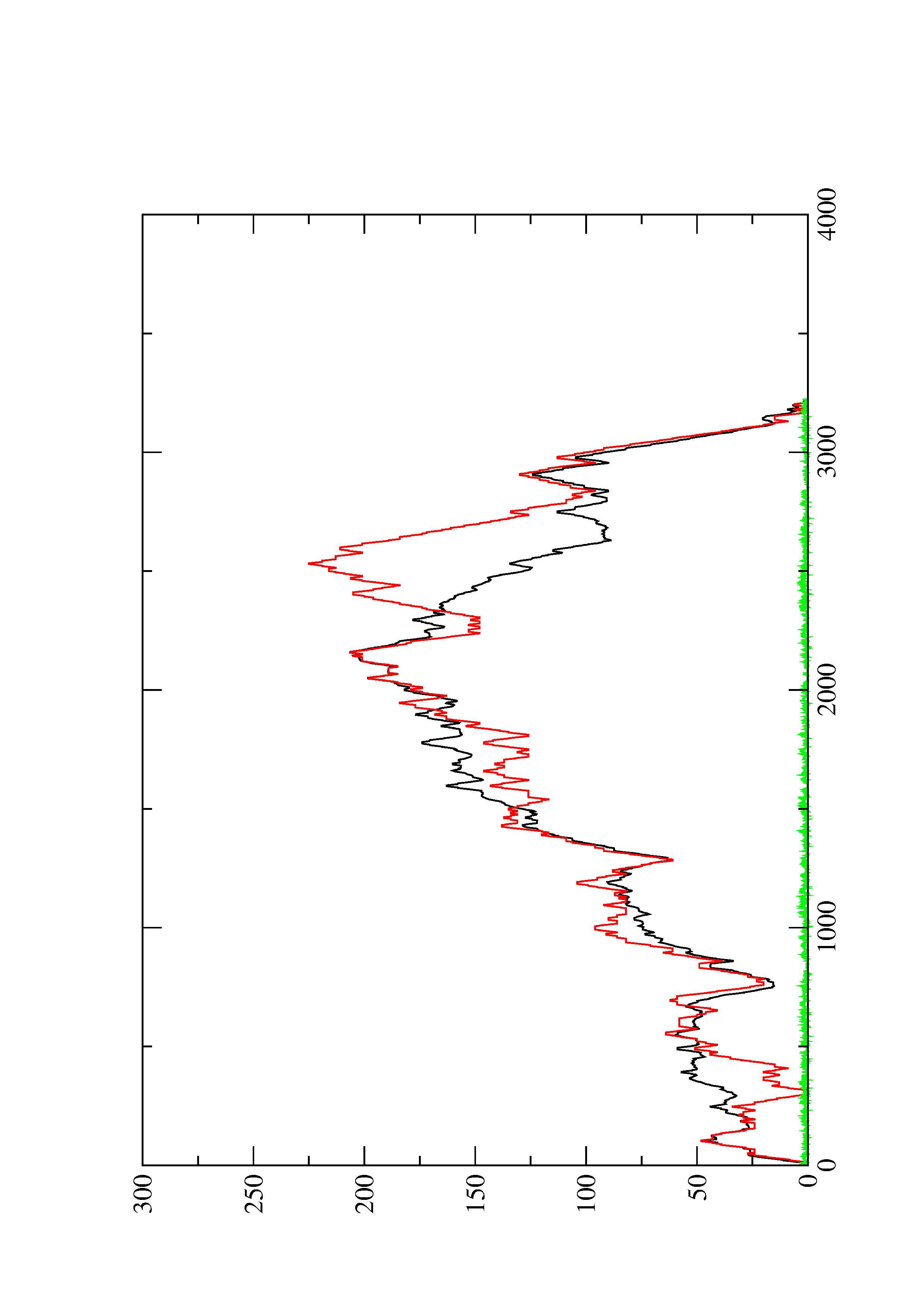
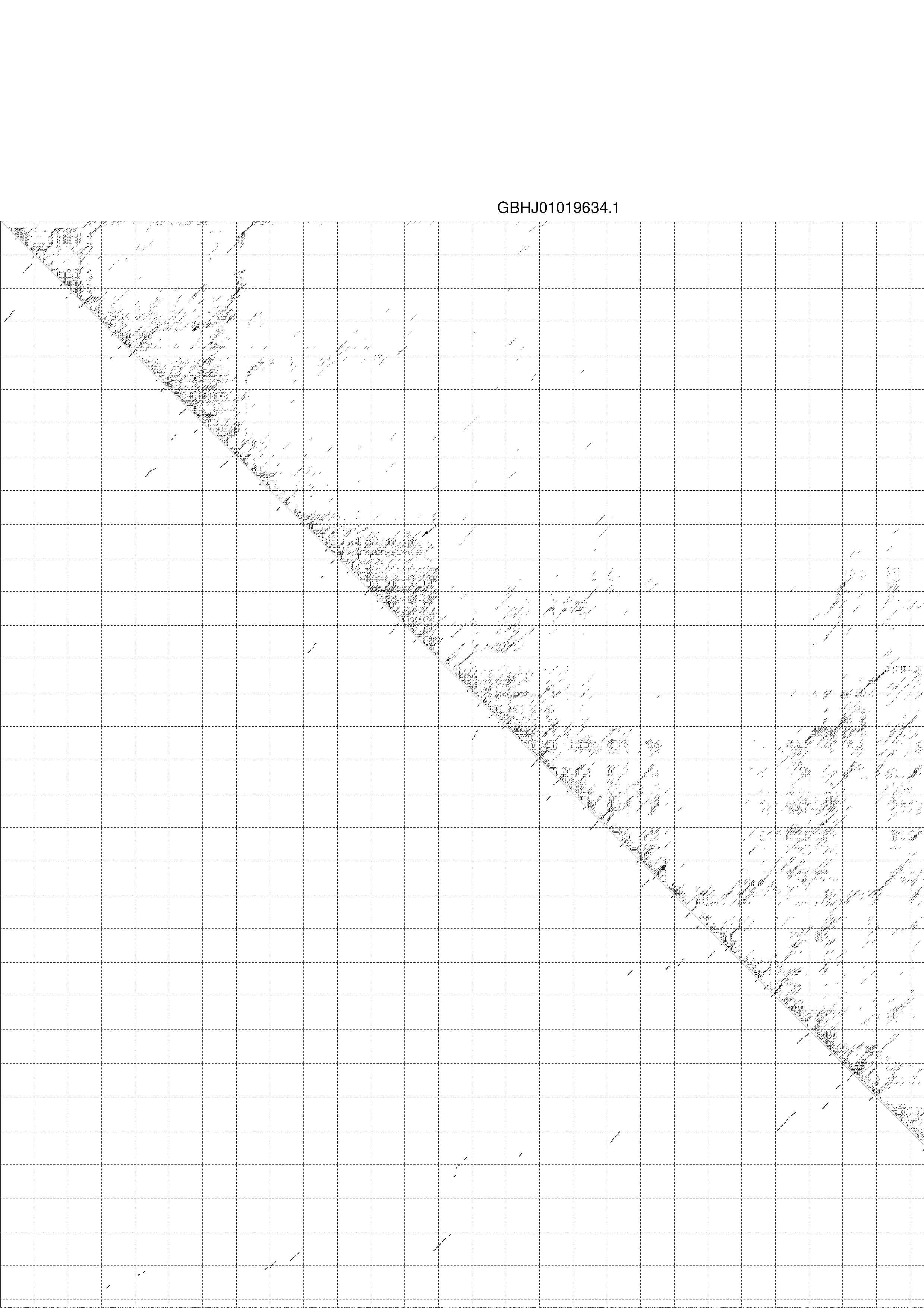
4. You may look at the dot plot containing the base pair probabilities [below]
III. Graphical output
1. A note on base-pairing probabilities:--
The structure below is colored by base-pairing probabilities. For unpaired regions the color denotes the probability of being unpaired. The reliability information of RNA secondary structure
plot in the form of color annotation, Positional entropy ranging from red (low entropy, well-defined) via green to blue and
violet (high entropy, ill-defined).
2. Here you find a mountain plot representation of the MFE structure, the thermodynamic ensemble of RNA structures, and the centroid structure. Additionally we present the positional entropy for each position. The resulting plot shows three curves, two mountain plots derived from the MFE structure (red) and the pairing probabilities (black) and a positional entropy curve (green). Well-defined regions are identified by low entropy.