lncRNA Structure Prediction
Species Name : Withania somnifera Sequence Id : GBHJ01020061.1
I. Result for minimum free energy prediction
The optimal secondary structure in dot-bracket notation with a minimum free energy of (-498.10) is given below.
1. Sequence-
UUAAUUUAAAGCUUUCAUUUCAUAAAACAAUAUAAGAAAACCUUUGAACAUAUACAAAGA
UGUAGAACUAUACUUAAGACAACUUGAGCUGAUUUCAGAACACAAACAUGAAUUAAAUAC
UCUAGUCUACGAAAUCUCUAACACAUGACGUCAGAAUAGAAAAUAUCAGGACAAGGCCCU
GGCGUACCCUAAAACAGAAAAAUACAUCAUAGAUACAAAAUGACCACGGACAAGAAGUGG
GGCUCACCAACUCCAACCGAAUACAAACUGAUAUGCUUAUCGAGGAGACCCAUUUUCUUG
GACACCUGAACCUACAUCAUGAGAUGCAACACCCCAACAUAAUAAAAAGGGAUGUAAAUA
CAUAUGAAAUUGUACUCGUAGAUAACAAAUUGCACUCGUACAGAAAGCAAAUAGAACACA
AUGUAUGGUAAAUAAUAAUCAUAAGAUAAUGAAAAGGGAUGUAAGUAUAUAUGAAAUUGU
ACUCGUACUUAAAGUAGAUAGAACACAAUGUAUGAUAAACAAUAAUCAUAAAGGAAUAUG
UGAAAUAAUGAAUGUAAAUGUAAGGAUCAUAAGGUAUCCUUUUCGGUUCGUUAUACAAAC
UUAUAUAUUAACUUUGUUUAAAAUAAUAUUUAAAAAGAAAAUCUCGUCAUAGUUUUUGUA
AUAACAUUUGAGGAUUUUAUCUACCAUAUUGGCUUUUUCCCUAGGCACGAUUGUUUUACU
AUUAUCUUAAUUUUAAUCUGCCACAUCCAGGUCCGCAGGCGGUCAUGAUGUCAGCAUGAA
AUUAUCGUUCAUUAUGUACCACGCUCUGGCCCUUCGUGCUUGGCAUGGUCAUUAUACCAU
UAUGAAUUGUUAUUUACCAUGUCUGGAGUCUUUGAGUUUGGCACAGUCGUAUCUCUUCUU
UUAAUCUUUUCAAGAGUGAUCAAUUCGUGUGGAUACUUUAGGCUACCUUAUUACUAUUUA
UUAUCUUACUCAUAUAGAUAAUCAUUUAACAUCAUGUCUUUAUUUGGGAGAAAUUUAUUC
AAGUGUGCGUUGGUAAACUUAGUUAUCUUCGGGAGAGAGAUCUUGAGCAUAUGUUAGUCA
UUCUUGGUUGAAGGAAAAUAGCACCGUUGUAAGAUACAUUAUGCACCAUCUAUGGUUACA
UUAUUUCAAGGGUCCCAUUCUUUUAUAUUCAUGAUACAUUAUUCAGUUUGAAAAAUAUUA
UACAUUUUCUUAGUCAUGUUCAUUAGUGUGAAGUAUAUUUAUAUGGAAGACAUAAAGAGU
UACAAAAGGGAGUCAUUUGUGAGCUUUCAUAGUCAUGGAUAUAACAACAAAGUAUUCUAU
UAACAUUAUACUCAUGGAGGGUACAUAUCGUCUCAUAUAUAGCAUGAUACCGAAGAGGGU
UCUUUAAGGUUAGGGGGCUUCGACCCUUCUUUGAACUUGUUACAGGAAAAGUACUUAAUU
UUUCAAGCAUUCAUCUUAAUUAUGAAGCUUAUAAAAGUUUACCAUUUUCUGAAUCGUAGC
GCAUUAGCCUCCUAUUGGAUAGAUAGCGUUCUUUAUUCGAAGACAUAACAUUAACGUGGA
CGGGUACCAUUUGAAAUAUUCUUAGACGUAGAUGAAAUCAUGUGAGGAGCUUAUGAACUU
GAUGUGGAGACAUAAUAAACAUAAAACGGUUAUAACUACAAAGUGUCCUAAAUACAUAGA
AGGCUAUUAGAUUAAGAUCUAUGGAGCAGUAAAUCUUCAAGUCUUAACAUAAUAGGCAUU
GAUAAAGAAGCAUCAAAUGAAGGGGUUCAUCUUAUCAAGAAUUGAAGUACUUUAAGGAAC
AUCAAUUUUUCAAACUCUUUAAGGCAAAGCAAACAUUAGUCAGAGGGACAUGCUAUAUCA
AAAAGAGAGGAAUUGCCUUACAUACCUCGAUUUCCGACCUUAAGCAUAUAAAAUUAGCAA
CUACUCCAUUGAGAUCUUAAUUUACAAGAAUAACGAGAAUAGGAAGUUAGAAUUAGCGAA
GAGAUUUGUGACUAUGCCACUUUGGCAAAUGGUCAAACAGAUGGUGAGAUAAAACACAAA
UUCUCCACCAAUGAUGUAACAAUAGACCACCUCUUCCUCCUUGUUAAAAACAACAUAAAA
ACUCUUCUCCGAGGGAUAACCCUCUGAGCAAUAAUACCACAAGAACACCUCAAACAAACC
UCAAUUUUUCCCUAAAGCUAACAAUAAUUCGUGCGCAUAGUUUUCUAUCAUCAUAUCAUG
AGACCCACAUCAAAAAUUUCAGAAAAUAUCAGAAACAAUCCAUACAUGAGUCUAGAAGAA
AAGUCUUACCUUAUAGAAUAAAAAUUCCAACAAUCAUCACCUGAUCCUUAGGUGAAGAAA
CCUCUUUGAAUCUCUUGAUGGUAGAUGGUUUAGAGCCGAAGUGAAUGGAGAGGAAAUAGC
UCAAAU
2. MFE structure-
...........(((((..(((((..................(((((........)))))..(((((.(((..((((((....))))))(((....)))........................)))))))).....((((((..(((..((((((...(((.....(((((.......))))).(((((((.....((...
........((((((((...............(((((.((((((((.....((.(((((((......(((((((((((((.((((((((....))))))))..............((((....))))..(((.(((.............))).)))...((((........))))((((.(((((((.((((((...((((
((.....(((((((((((..((((((((((((((((((((((((.(((((((..(((((...((((..(((((..(((((..((((((.....................))))))...)))))....((((((.(((((((((.(((((((((..(((....(((((........))))))))..)))))))))......
)))))))))..))))))(((((......)))))..(((.((..((.(((..(((........)))...))).))..)).)))..)))))..))))..))))).((((.(((((....................))))))))).(((((((((.((.(((.(((((.(((((..((((((............))))))...
))).)))))))))))).)))))).))).)))))))....))))).)))))))))))))))....(((((((..((((((.(((((((.((..(((((..............)))))..))..))).)))).)).)))).))))).))............(((((((.........)))))))......))))..))).))
))))))(((........)))...))))))...))))..)).))))))).))))(((....))))))))))..)))))).....))))))).)).......(((....((((...))))...)))((((....)))).............)))))))))))))(((((((((((((......((((...))))..(((((.
((((((..................)))))).))))).....((((((((.((((((((.((........)).)).))))))..))))))))))))))))))))).......))).)))))..........)).....))))))).((((((((.......)).)))))).(((((((((((((.(((((((((((((((.
...........((..((((...(((((((.(((...(((((((.((((.(((((.......)))))))))..))))))))))..)))))))...)).))..))..)))))))))(((((((((........)))))))))((((....((((.((((....(((((..........)))))....)))).))))...(((
((((.(((..........(((((((((.(((((.............................)))))........(((((((((((((((((((((((((((((((.(((...(((((((((((.........)))).))))...)))........(((((....)))))......(((((((((.((.((....)).))
.)))))))))....((((((..(((.((.......)).)))))))))..(((((...((..(((..((((.((........)).))))..)))..)).....)))))............))).))))))..))))))))))))).........((.((((.....(((((..((((.((((((.(((((((((((((((.
..)))))..))))).......((((((((............))).)))))(((.((((((((.((...........))..)))))....))).)))..........(((.(((((....))))).))).........................))))).))).....))).))))..)))))....)))).))...))))
))))))))..((((...))))...)))))))))...((((.((.....)).))))...))).)))))))))))..((((....)))))))))).)))))..............((.((((((((...)))))))).))..))))))))..))).))))))....)))..)))))).....)))))..)))))........
......
You can download the minimum free energy (MFE) structure in
II. Results for thermodynamic ensemble prediction
1. The free energy of the thermodynamic ensemble is [-542.80] kcal/mol.
2. The frequency of mfe structure in ensemble 3.1622e-32.
3. The ensemble diversity 626.28.
4. You may look at the dot plot containing the base pair probabilities [below]

III. Graphical output
1. A note on base-pairing probabilities:--
The structure below is colored by base-pairing probabilities. For unpaired regions the color denotes the probability of being unpaired. The reliability information of RNA secondary structure
plot in the form of color annotation, Positional entropy ranging from red (low entropy, well-defined) via green to blue and
violet (high entropy, ill-defined).
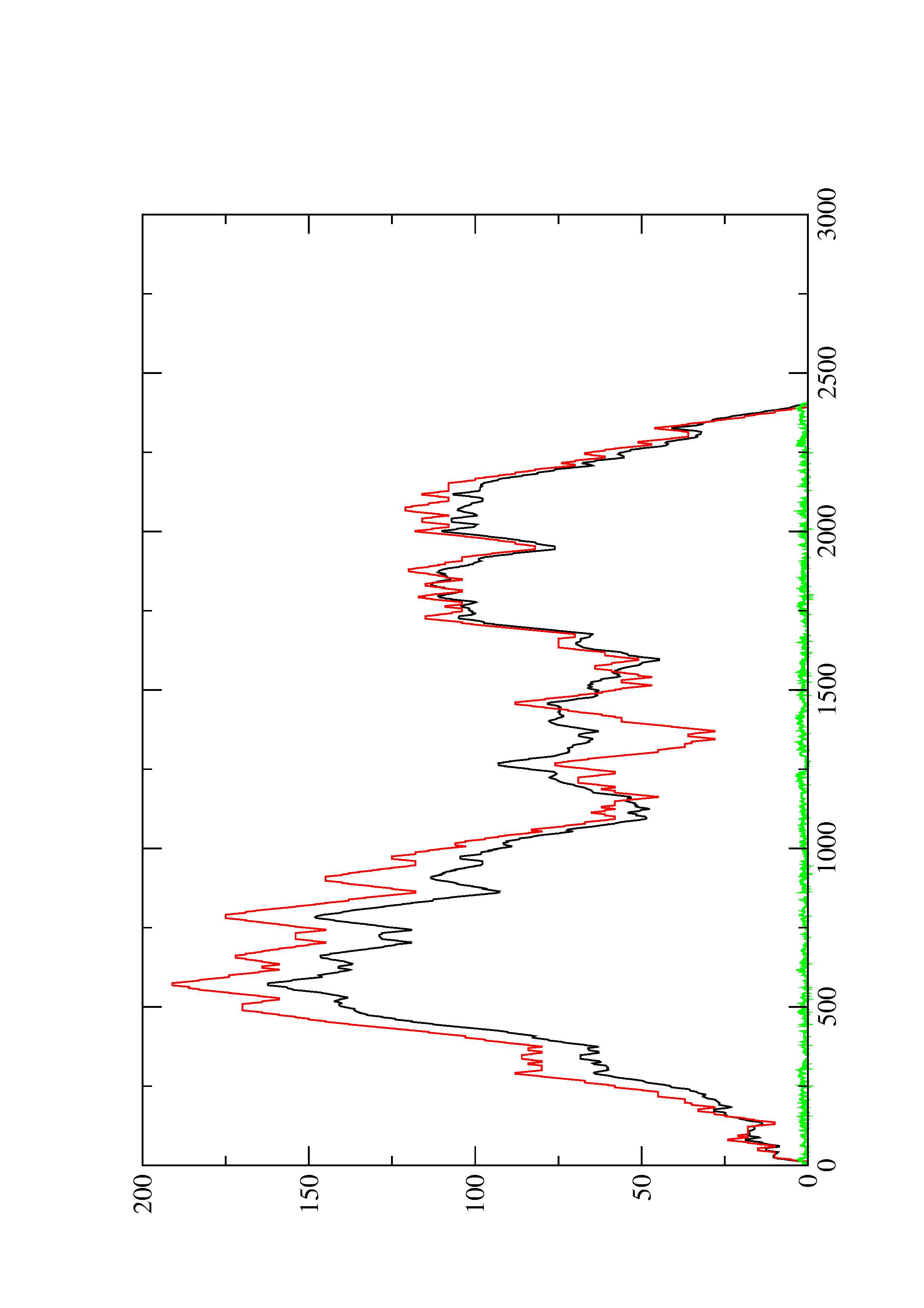
2. Here you find a mountain plot representation of the MFE structure, the thermodynamic ensemble of RNA structures, and the centroid structure. Additionally we present the positional entropy for each position. The resulting plot shows three curves, two mountain plots derived from the MFE structure (red) and the pairing probabilities (black) and a positional entropy curve (green). Well-defined regions are identified by low entropy.