lncRNA Structure Prediction
Species Name : Withania somnifera Sequence Id : GBHJ01020195.1
I. Result for minimum free energy prediction
The optimal secondary structure in dot-bracket notation with a minimum free energy of (-534.30) is given below.
1. Sequence-
GUUCUUGAUCUUCUUCCAUUUGGAUAGUAGAUGGCAUCUUUAUGCAUCUAGUUAAUCCUU
GAAUCUUCUUGAAUUUCAGAGAGAUAUUUUGGUGUAUUAGAAGCUACCAUUGACAAUUUU
AGCUAUGGUUUCCAUGCUAUCCAAAUUAUUUACGAUGAAAACGGAAUAGUAGAAAGUGGU
AGUUCUUUUAUAUGCAAGUAACAUACGUGAGGUUUGUUCCAUAUGAUCACCCGGACCUUG
UCUAGGAAACUUCUACCUGCAUUUUGCCUUAGUUUUAGUCUUUUUGCUUCAUCAACAGAU
AGAAAUGCCAGUUCACCAUGGAAAAAGGAAAUCAAAAUAUAAGGUGCUUAAAUAAUCCUU
CUUCAUGUUCUUUGUUUCUUGUCAAGAAAGAACAAAAAUCCAUGCUCAGUAGACCUAAUA
GGGGAAUUUCCUUCUGUUUUAUUAUUUAUUUUAGAUUCAUCAGAAGAGAUGGUAUCCCAU
UUUUUCAUAAUGAAAAAAGAAAACUAAGUAUGCGAUCACCAAUGCGAUAUAUCUUUCUCC
UAAAGGCUGGAAUGAUCUUGUUAUAUGCAUUGAAUUCAUCAGAUCUUUUCUUAUGACCUA
UGUAGCAUGGGUUAUUUUAUGAAAACUGCAGUUGGUAUGAUAGGUUUGUAACUAUAGGAA
UUAUAGGGAUAUGAUGAAGUUAUGCACGCAUUUAUUCAUUGUUUUAUAUUGCGCUGUAUA
UUUUCAGUCAAAAAGAAUAUUGAGUACUUUAUAACCUAUGGCAAACAAUGUAAGAAGUGA
GCAUUAGGAUUGCUGAGUUCAGUGCAAUUAUACCAAUAGGCAAAGUGACAAUUUACAGUU
UGGAUGAGUUGCCUCUAUCUUCCUCUUUGUGGCUUUUCAUCACAAUCUCUGGCAUGCUAU
GCAAUGCUACUGAGUCCUCUCUAUUAGUUCAUCUAUGCUUCCCAAAUUUUUUUUAAAUGA
GAAAUUGUGAGCUGCAUAACCAUUGUACCUGUUAGAAGCAGUCCUAUUCUUUUUUCAGAU
CCUUUGAAAACUUUACUGCUAAACAUUAGUAGUUUUUUGAUAACCGUGGUCUCCGUGCAA
GCUUGCGCGCAUCUAAAUCCUCCCACCAGCAACAAGUACCAUCACCAUGUAACUCUGUCC
ACCAAGUCUAGGAGAGAUGGGAAGAAAUCUUCUACUAAUGUUUUUUUUAACCUGAAACCU
CAUAGUCCUCAACCACUUCAUCUACUUUAAACAGUAGAAUUUUUUUGGUGUUGUAUCUCA
AUUGUUUGUCCACUUAAAAUUGUUUUAUCCAUUUGCAUCGGUAAUCAUCAAAUGGAACUG
UGAAGAGCCAUCCUAUAUGCCUUUUAGUUUGAGAAGUGCUUCGAAGUUCGAACAUCCUGC
ACAACAUCACGUGAUUGAGAUAUUGUAUUGUCACUUGAAAAUUUGUGUGGCAGAUACAUC
CUAAGGCAGAUAUCAUUUGACCAAGGAGUAACUGAAUAGAUCCCAACUGCUAUUAUCUUU
UUGCCAUACAUUGGUUUAGCUGGGGUAAAACACUUUGAGAUCCAAAAGGAGGAGAAUCAA
UGUUGUGAAACUAUGAAUGUAAAAAAGAGCAUCUUUAUUAAUUCUCAUUUUGAUGCAGUG
UAAUGCGUUCACAACUACGUGAACAUCUAGUUUCUCAAAGAAAGAUUUCCCUAUGACUUG
AGUGGUUACAUAUUUUUAUUUAUCAGAGUCAACCUCUUCUUUGGUUUAUAGCGAAAUAAU
GCCUUGUAAUUAUGCAAACAAUGGUGUGGGUUCUAUAUUGUGAGUUAUGCCACUAGGUAC
AUUGUUCUCCAUUGAACAACAAAAAGUCACAAUUAGUUUCCCUAUACUGUUCCUUAACAA
AUAUUGUAAAAUUUGCAUUUUUAUCCCGUUCUUAGUAACCAAGGGCAAAACCAAGUGGGU
GGUAGGGGGUUCGGCGAAUCCCCUUGGCUUCACGUGUUUACUUUUUUAAUAUUUUGAACC
CCCUCAGUGAAAUUCUGGUUCCUCCACUGGUAGUAACGUCACCAAGUUGAUGUUUUACGG
UUCUUGUAGUGUUAUCAAAGGCAUGCUUAAAGAACACUUAAGCCCUAAAGCGAGGCUCAA
AACGUUUUGAGUGCUUUGCGUCGCUUUAUGUGCGCGUCAUUGUCGUUAUCAAGUUUCUAA
GGCAUAUUUUUCCUUGUCAAUGAGUCUCCUGAAAAGGCGAUACUAAACAAUUGACAUAUC
ACUUUAUCAUAAUUGUUUUUCAAUUUGUUUGU
2. MFE structure-
((((..((.....(((.....((((..((((((.(((....)))))))))....))))..)))...))..))))(((((.(((((...(((((((.(((((((((.....(((((((..((((.((((...)))))))).....(((((((.((.......))))))))).......((..((((....((((..(((((
(.((((((((((((...........(((((((((((...))))(((.........)))(((...)))(((.(((((.(((((((((((.....(((.(((((((((((.((((.(((((((.........(((((((......(((((((.......((((((((((((((((.((.......)).)))))))...((((
((((((..((((....((((((((......))))))))....((((((((((...(((((..((((((((((....))))))))))...))))).....)))).)))))).........((((((.(((((....(((((........)))..))......)))))))))))...((((.(((.....))).)))).)))
)..))))))))))...(((((..(((....)))..)))))...(((((...(((((((..((((((((.(((..(((((.((((((((((...................)))).)))))).))))).................))))))))))))))))))))))).))).)))))))))))))((((((((((.....(
((((((((....((....))((((.((........)).))))((((((..(((.................)))..))))))........((((((((...)).))))))..(((....)))(((((((((((............((((((((......)))))))))))))))).)))...)))))).))).....))))
)))))).....((((((((...))))))))....((((((((...)))))))).)))))))..)))))))...((((((....))))))......................((((((..(((....)))..))).)))((((((((((((((.(((((.(((((....)))))....(((((((((((.((........(
((((((...((((.......((((((......))))))((((((((....(((.(((((((.(((((....................((((((((.((........))))))))))((((.((((.((..........)))))))))).......(((.(((((...)))))))).((((((((..(((.(((((.....
.........))))).)))....))))))))(((.(((.((((..(((((((...))))).)).))))))).))).....((((((...((((..........((((.....)))).)))))))))).)))))..))))))).)))....))))))))....))))....)))))))..)).)))))))))))....((((
(........)))))(((......)))((((((......))))))......(((((...))))).))))))))).)))))).))))...(((((((((((..(((((((..........)))))))..)))..))))).)))..(((((....)))))..(((((((((((..(((.(((((((.((.((((....)))).
.((((((......))))))....)).))))))))))....))))))))))).....((((...))))..)))).)))))))))))..)))...)))))..))))))))))).))).)))))))(((((((((...))))))))).)))))))))))))))))).....))))...))))..)).(((((...........
....)))))((((.(((((((......)))))))))))((..((((.((((((....(((.....)))....)))))))))).)).(((((...(((((((....))))))))))))(((.(((.....))))))...)))))))......))))))))).))))............)))...)))))))))).((((((
....(((((((((..(((......)))..)))))))))......))))))..
You can download the minimum free energy (MFE) structure in
II. Results for thermodynamic ensemble prediction
1. The free energy of the thermodynamic ensemble is [-576.86] kcal/mol.
2. The frequency of mfe structure in ensemble 1.02409e-30.
3. The ensemble diversity 631.97.
4. You may look at the dot plot containing the base pair probabilities [below]

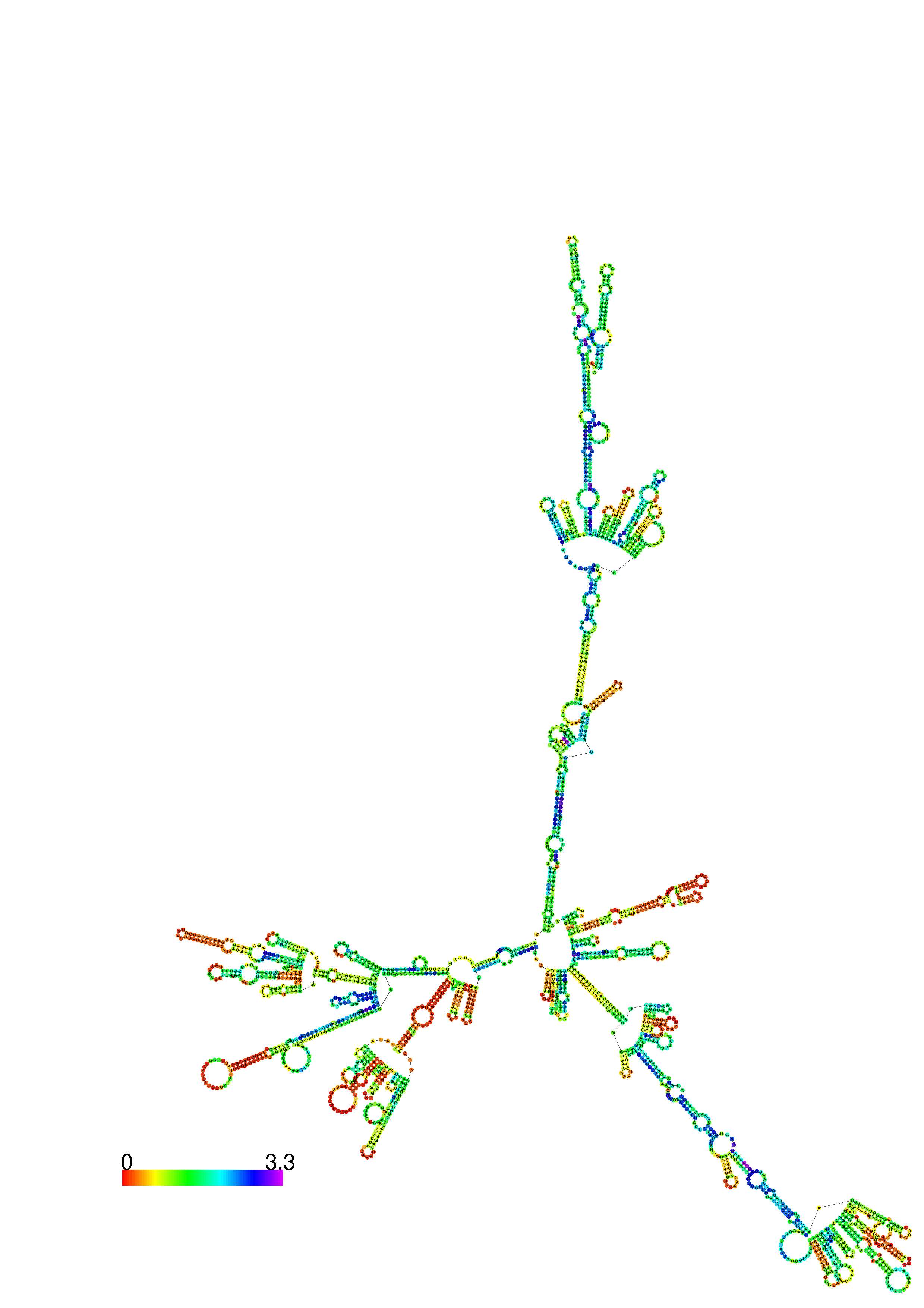
III. Graphical output
1. A note on base-pairing probabilities:--
The structure below is colored by base-pairing probabilities. For unpaired regions the color denotes the probability of being unpaired. The reliability information of RNA secondary structure
plot in the form of color annotation, Positional entropy ranging from red (low entropy, well-defined) via green to blue and
violet (high entropy, ill-defined).
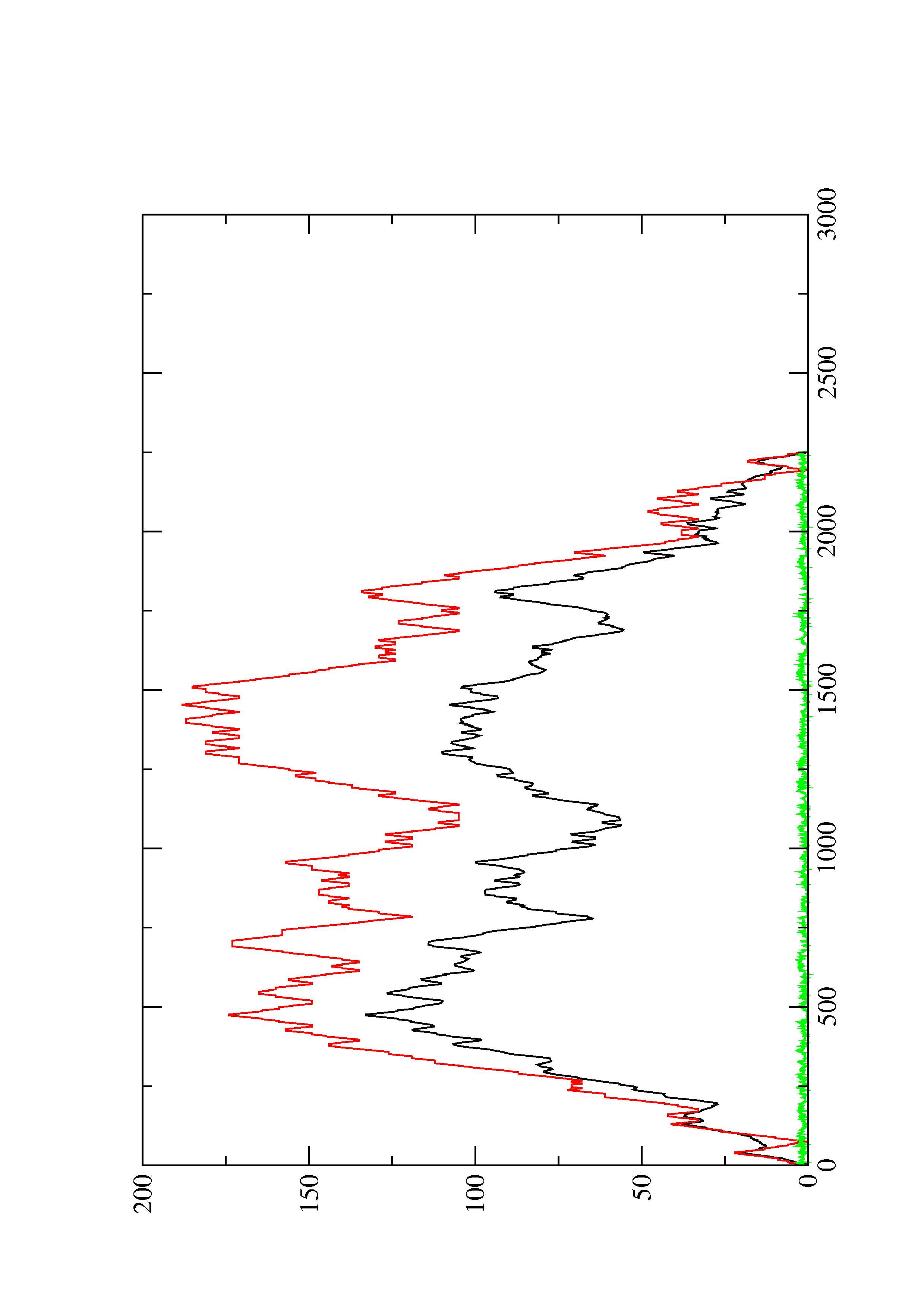
2. Here you find a mountain plot representation of the MFE structure, the thermodynamic ensemble of RNA structures, and the centroid structure. Additionally we present the positional entropy for each position. The resulting plot shows three curves, two mountain plots derived from the MFE structure (red) and the pairing probabilities (black) and a positional entropy curve (green). Well-defined regions are identified by low entropy.