lncRNA Structure Prediction
Species Name : Withania somnifera Sequence Id : GBHJ01021654.1
I. Result for minimum free energy prediction
The optimal secondary structure in dot-bracket notation with a minimum free energy of (-590.80) is given below.
1. Sequence-
AUUAAGGUAGGGUGAUAGAGAUAUAAUCUUGAGUUGCAAUCGAUCCAAAGUUACUUGGAC
CUGGUUCUCCAUAUUUGUUCACCUGGGAAAGAACCGAACCCGAUACAAUUACUUUGGUAC
CUGCCCUAAUUCUAACGGAAUUUGGCCUCCAGCCCAUGAAUGAAAAAAAAAAAAAAGAUU
GCUGCAUUCCCUCAAAGGGUUUGGACCAAUUUAUCCUUCGCCUUUUUCUCCCGUCUUCUG
UAUGUAAGGACUUGGGAUGGUUUUUUCGACUCAAUUUUGGCUUUGGGCCCAAGCGAAGAU
GAGUCCAAUGGACUCGGAAAAGAGGUUCAUGCAAAUAAAAUGAUAAGUAUUAGUGUCAAC
UCAAUCCGUAAUAAUAAAGAACCGAAAAUAUAAAGUGAUCAAAAGAAAAGGAUUUCCGGC
AACGAAGUGGGCAGGCAACGUUAGAUAUUAUUCUUAUGAAGAAGUUGUUCAUGUUUACUG
UAAACUACUCAAAGCCAUAGAACUUCUAAUUACCGCACGAGUGAAUACUUAUUUCUAAUA
GCUCUUACCAAUUCAAAUAGAGGACAUUAGACUUACAAACUCAAAUUCACAACUGGAAGU
AGAUGAUAUGAGAAGAAUGAGAGAAUUGACAUCAUCAUACAUACUGGAAUCAACCUAAGU
UACCAAGCCUUAUUCUAUCAUGUAUGAAGAAGGAAAGGCAAAACACAUCUUUGUCAAGCU
UCAAGGACUCCAACUUUGAACACAAACAGGGGGGUUAAGUUAGCAUGUUUCACUUUGAAG
AAAGGAACCAGCCUACUCAAUCUAGGUGGUAUUAAACAGACAAUCAGCAUACUUCUUCAA
AAAAAUAGAUGCAAGACAAAGUGAAGUGGGGAAACAACUCAUGAAGUUCUAAUCUUUUCG
ACAUGCGGACUAGUAACUAAAGAAAAUGAGGACAAGCAAAGAAAAUAAAGAAAAAUUACU
GCCUGUCAAUUCGUCAUACCAAGCCAUUUAAAACAGAUUACAUGUACUAUUUUCUUGUUA
GAAAGCCACACAAAUAAACACUUGAUGGGAUCAUGUUGAAAGAAAAUGAACAAGGUUAAC
UAUUAAGCCUAUUUGUUAGCUAAUAGACAAUGCAACUUGUUGGUUGUAGUUUAGUAGACC
CUAAGCAUACUUAUAGGUGAGCAAGUACAAAAGUAAUCAAUGAAACAAGGCACAAAGAUU
CAGGCUAACACAAGUUAAAUAAGGCUUGACUGGAUUACUAUUAAUCACCAGAGAGGCCAU
UUCACCAGGUUCUAAACCUUACUGAAAAUAUUUUGCUGGUGAGCAGAUUUUAAGAGAUUA
AAUCACCUUAUUGACCUUCUAAUUGAACCAGAACAGGAAUUGUUACAGGAUCUUUACUAG
ACAAGCAAGUAGUUACUAGGAAGGUAUCCUAGACUGCUUACAAGACCUUCAUGAGUCUAC
AGUGAUUCAUGUUAGACUCCCAUAUCAAAAAGACCAUCCCAGAUCCAAAACCAAAUUAUA
CACAACAUUAGUUAUACAGACUUGUGUUAUUAAGCACUAAGACUUAGGUACCUAAUCAAG
UUCAAACCUAUCAAACAACCACAGAUAUCAAGAUCCUCAGAACCUCUCACCCCUCCCUUA
GGACAUGAAUAAUUAAGGAGACAACAAAUGCAGGACUGGAGAAUGUAAUAACAACAAUAA
AAGGGAACAACUCACAAAUGAACAUUAAAUAACAAAUAGGUUCGCAUACUUUCUUCAUAC
UGGUUUAACCUAGAGACUUGCUCAUACAUAUAAAUCAAGUAAUAUCAUCUUUAUCUACUA
CAGGGUCACUAUAUAAGACAAGAAGCAUGCACAAGAAAUCACUUACUUUGAAUCUUUAUG
GAUUAGCUCUAAGUCCAGAACCACACAUCAGCUUAGCUACCACACCUAUCUCCAUGCAAC
CAAGCACCCUUUUCCUAUGCAGUUAGAUUCUAAUGCAUCAAGGGUGUUGCAUGUAAAUCA
UGUAUAACUCAGCCAAACCACAACAAGGUCCCGCUAAACUAGCAUUAACAGCAAGCGUUA
UGAAGGUUUUCAUUCAUUAGGCUUUAGAAAUAGGCAUUUAAAUUACUCAAAUUAGCUAAA
UCUGAGUUGAUCAAACUUGAAUAUGCAUACAAAGCUCCAAACACCGUGCAAAUACUCUAC
AUGUUUUUAAAAGGCUCAAACUUUGGCACAAGAGAAAGCAAAAUUCAAAACAGGCAAGAA
UUUUAACAUGCUGCAUCUGUUGAUUGUAUAACCUUAACUUAGCAUUAUGUGGAUACUGCA
CCCACAUAAUUGGACACAAUAUCCAAUAUUUUAAUAGGUCAUCUAAGUGAGAUCUGCGCA
UUUUAUUACUCAAUUAUAAGUCCCGUCUCUUGAACCAGAUUCUAAGCCCCCACAGGAAUU
AACCAGAAAAACCAAAGUUCAUCAAGAAGAUCUUUUGCUGCCGGUAAGCUAAUAGGGAGG
AUAGUGAAAUACAAAAAUUACUAAGGUUCCAACAAGUCUAAUUAAAGGCUUUCCAAGAAG
GAACUUGAGCACCGAGUUGACCUGGAACUUAAAAAGAAUGGUGACUUGGCAGCAUUCACA
GAUUCCUACAAUAGCACAUAGAUUGGGAUCACUAGAGCACAUUCUUGACGAACUAGACUA
ACUUUAUGGACCAUAAUCAAAACACAUUAGACAUAUAUCACCACAGCUUUAGGGAACAAU
AAAAAACAGAAACUUUUUAGGCAGUGAACAGGCAGUCUCCAAUCAAUCCUAUUCACUUAA
GCAGAAUGCACAUGCUCUUAAAUCUCAAAUGCUAUGCAAAUGCACACAGAUCAACUGAUU
CUACCUCAAGACUCAUGCUUUAACAUAAAAAUAAAGGGAAAAAUUAGUUAAGACCUUGUA
CCAAGCAGGACUUGUAGGCUUAACAUUAA
2. MFE structure-
.....((((((((((((((((((((..(((((((.((((((..(((((......)))))..(((....)))..((((((((..((((...((.(((((.(((.((.(((((....(((....))).))))).)).)))..)))))..))...))))))))))))............)))))).)).((((((.(((((((
..(((........)))...)))))))(((((.(((..((((((....(((.(((((((((((((((.((((((.((((.(((.((....))))).))))))))))..((((((((......)).))))))..........(((((........))))).....((((((((((((..................(((((((
........((((.(((.(((...((.((((((((..(((((..((((...(((((...)))))..)))).)))))..))))...))))))...)))...))).))))......(((....)))..........((((((..((((.............))))...))))))........((((...(((...(((....)
)).)))..))))....((((..((((((((.((.((((((((..((((((....((.........))....))))))..)))))))).)).....(((((.......(((((((....(((((..((((((...((((....))))....)))))).((((.(((((((((((((((.........((((.((((.....
..))))))))................((((.((.............))))))....)))))))).(((((.......)))))((((.........))))))))))).))))...............)))))....))))))).................))))))))))))).)))).....(((..........(((((
(.((((((...((((....))))(((...(((((((....((((((((((((.((((...........)))).))))..))))))))..)))))))..)))..(((((..(((((((....))))))))))))((..(((.((((....))))..)))..)).))))))...))))))..........)))........(
(((((((((....)))......)))))))...))))))).....(((((((((((....(((((..(((((...)))))...)))))...)))).)))))))...(((((.........))))).(((..(((...(((.....))).)))...))).)))))))).))))...............(((((((..(((((
(.....))))))))))))).......((.((((((((......))))))))..))............))))))))))))))).))).................(((((.((((.....)))).)))))............(((((.(((.....))))))))......((((...........)))).............
............((((.((((...........)))))))).......))))))))).))))).(((....)))........))))))....(((....)))...............((((((.....(((...........)))..))))))(((.....)))...........)))))...))).))))))))).))).
....(((.........)))....(((((.((.((((..(((.......))).))))..)).))..)))((((((...((........)).)))))))))))...(((((.(((.(((......((((((((.....(((((.((((...))))))))).))))))))((((((.....))))))................
..((((((((..((((...)))).......(((((((((.(((.(((((.(((.((((..((((.((.((((((.(((......................((((.((((((....((((....(((((.................)))))......................((((.....(((((..(((((((.((((
((((((...........)))))).....)))).....(((((((((........))).)))))).(((((((.........)))))))((((((.......)))))).......((((((.((.....))))))))((...(((((.........)))))...)))))))))..))))).....))))....)))).)))
)))))))........(((((.(((((((..(((..((...(((((..((((.((((((...((((((..........))))))((((((((..((((((......))))))(((.....)))(((((.....))))).....)))))))).....(((((.((......)).))))).....))))))...)))).....
.)))))...))..))).....))))))).)))))(((((..(((((((...))))).........................(((.((((.(((...................)))...)))).)))...))..)))))......)))))))))..)).))))..))))...))).....)))))))))))))).)))...
............((((((((...(((........(((......)))........)))....))))))))..)))))))).....))))))...)))))...........
You can download the minimum free energy (MFE) structure in
II. Results for thermodynamic ensemble prediction
1. The free energy of the thermodynamic ensemble is [-648.28] kcal/mol.
2. The frequency of mfe structure in ensemble 3.13765e-41.
3. The ensemble diversity 704.87.
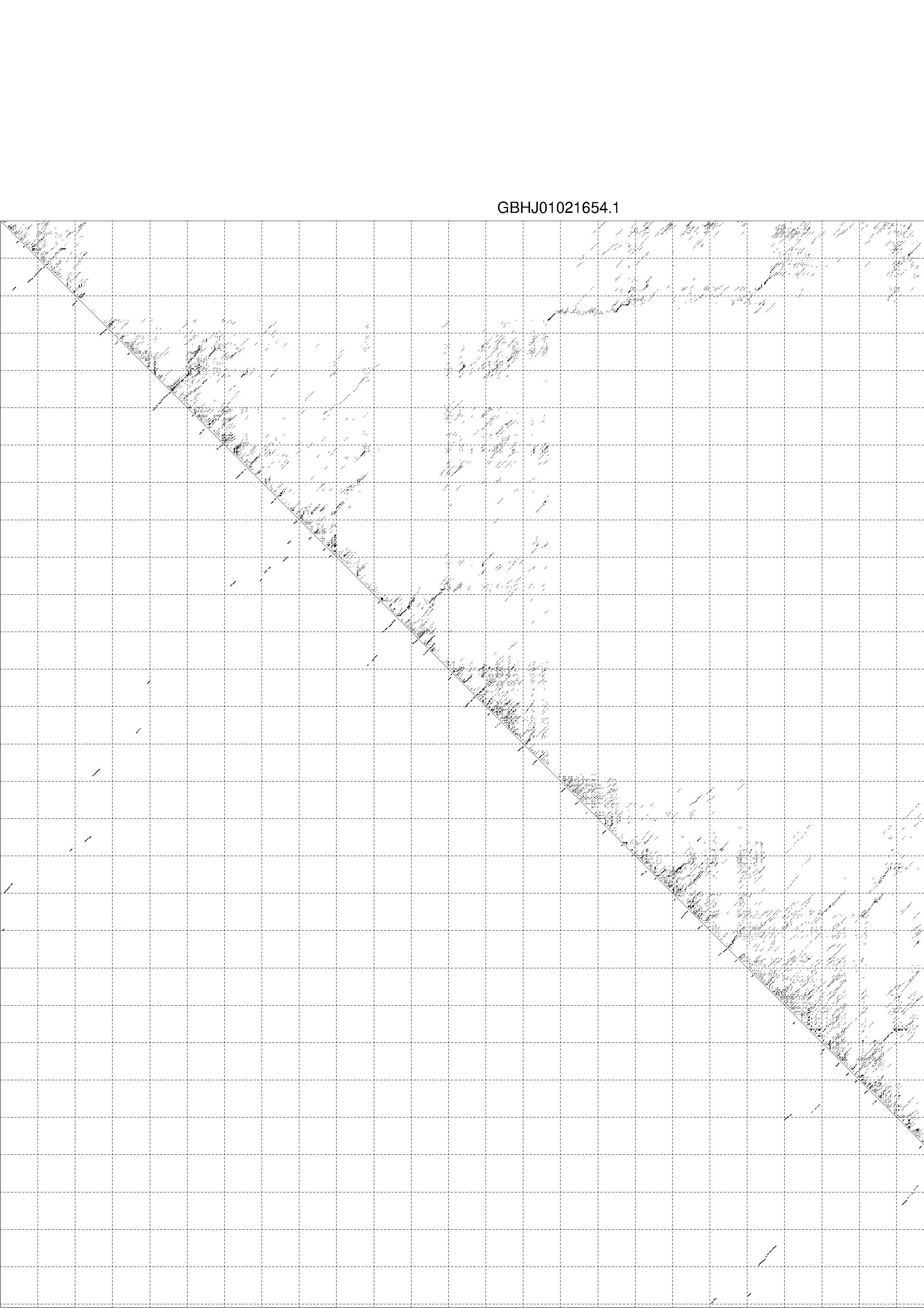
4. You may look at the dot plot containing the base pair probabilities [below]
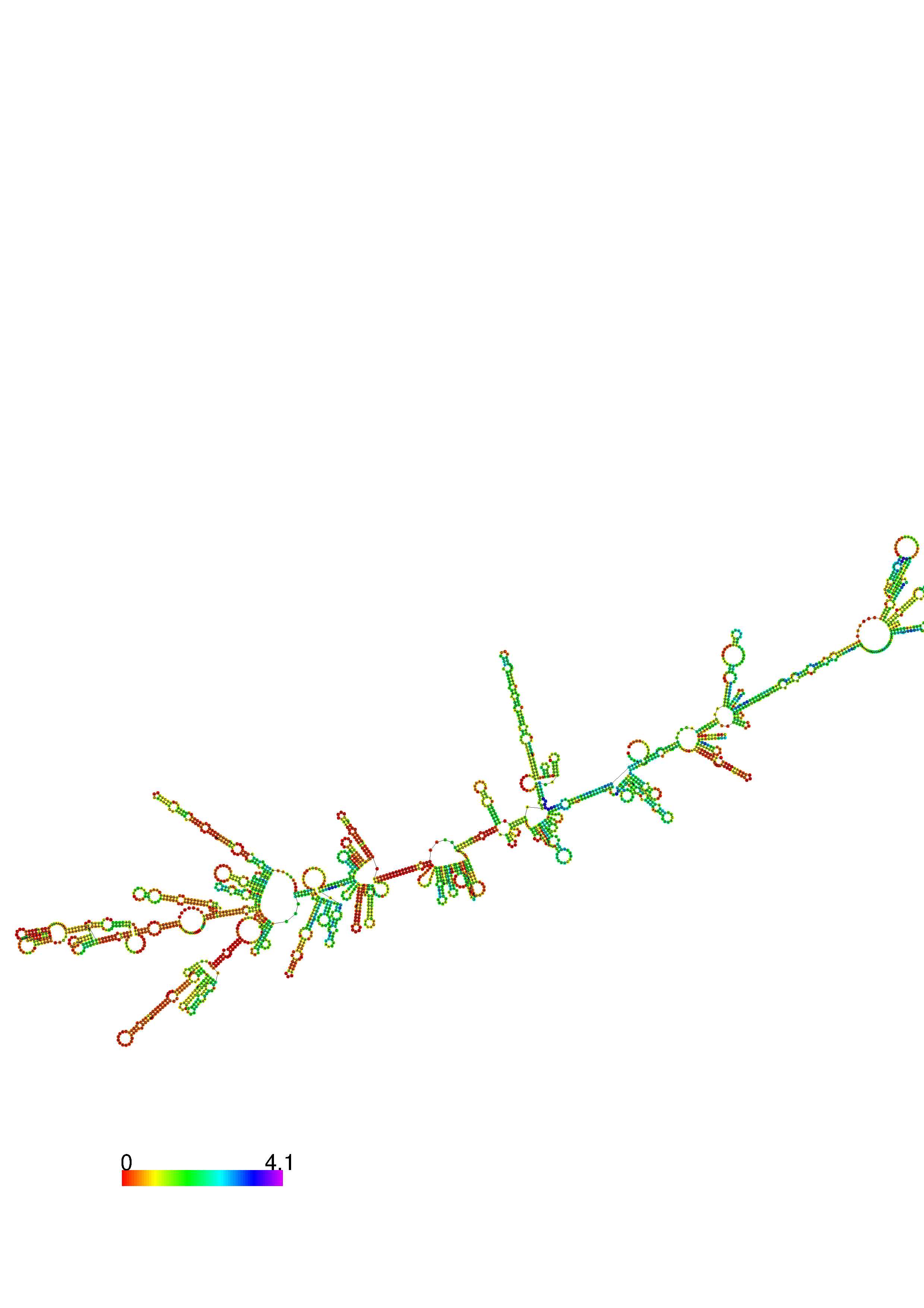
III. Graphical output
1. A note on base-pairing probabilities:--
The structure below is colored by base-pairing probabilities. For unpaired regions the color denotes the probability of being unpaired. The reliability information of RNA secondary structure
plot in the form of color annotation, Positional entropy ranging from red (low entropy, well-defined) via green to blue and
violet (high entropy, ill-defined).
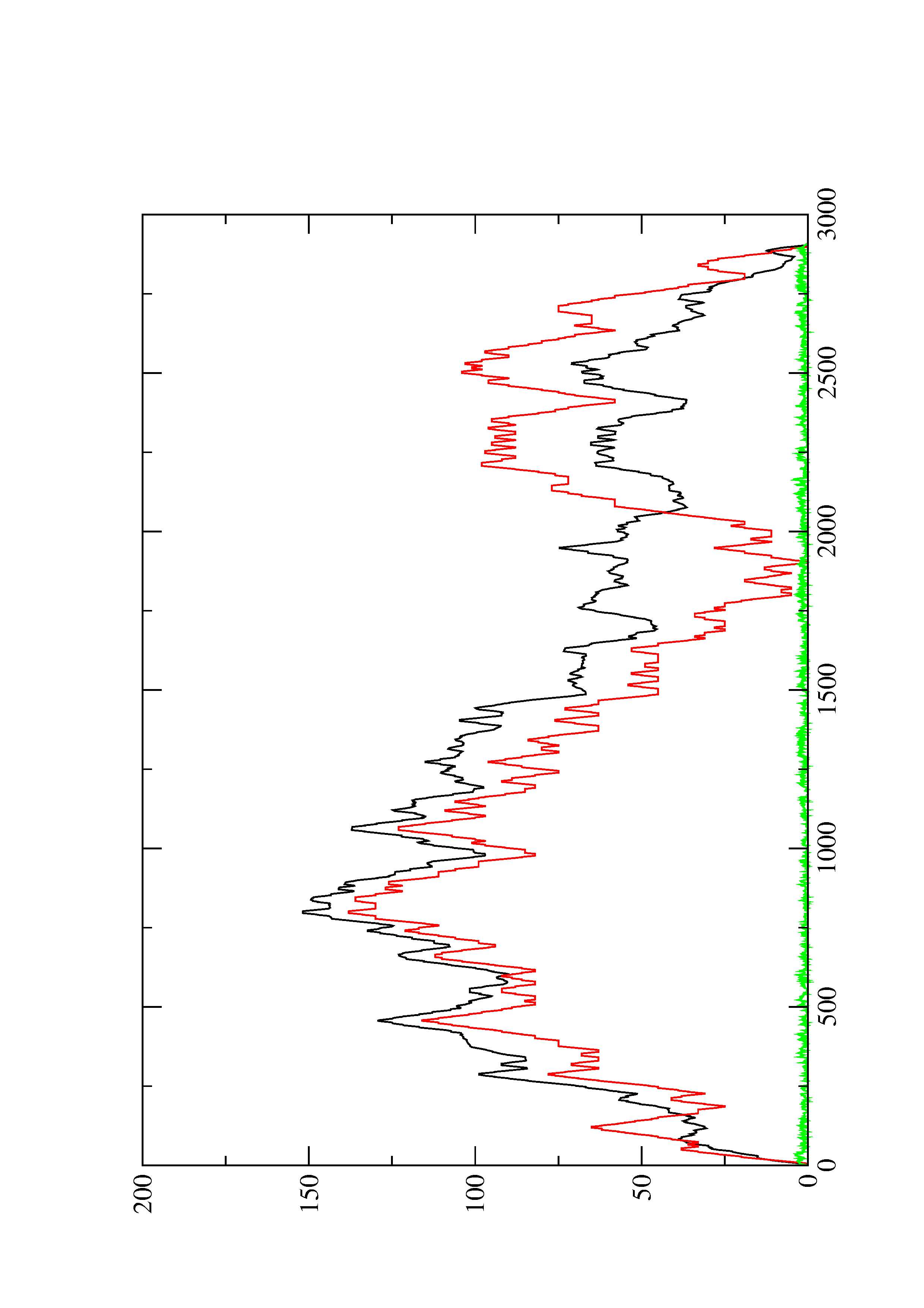
2. Here you find a mountain plot representation of the MFE structure, the thermodynamic ensemble of RNA structures, and the centroid structure. Additionally we present the positional entropy for each position. The resulting plot shows three curves, two mountain plots derived from the MFE structure (red) and the pairing probabilities (black) and a positional entropy curve (green). Well-defined regions are identified by low entropy.