lncRNA Structure Prediction
Species Name : Withania somnifera Sequence Id : GBHJ01021701.1
I. Result for minimum free energy prediction
The optimal secondary structure in dot-bracket notation with a minimum free energy of (-549.30) is given below.
1. Sequence-
CUAAGAUCUGAUACCAAAUGAUGCAGUUAGAUAUACAAAACGCACGAAACACAAGGGAGA
AAGAAAUAAAGAAAAAAGGAUAUUCGAAAACUUUAACCCACGGUUUAUGAACAACAAGGG
ACUGUUACCCUCACCUUUCUAUCGGACUUAUGAGUUGAUGACAAAUCUCAAACACACAAU
CUACGGAAAACCAAAUUGAUUCCAGAAAAUAGAAAAUCCUUCGAUUGAUGUUAAAAAGAA
GUUACGAUUGCAACUAAGGGAAUCCACCCUCAAUCACACACUAAUUUACGCUAGAUUACG
ACUAAAAGCUAUUCUAGGCUAAGAAUUAAUCUCACAAUGGAGUUCAAAUAAUUUCAUCAA
UGUCUUUUGAAAAUUACUACAAACUCUAGUAUUUAUAGUUAUUACAAGUUUGUAAUAAAG
GCACUUUGUCCCCCAAAGUCUUGGCAUCUUCACAACAGCCCCCAAAGUUCUGGGAUCUUC
AAUUAGGCCCCUCAAGUUCUGGAAUCUUCACAUCUUGCAACUCACAAAAUUUCCAUAGGU
UGAAUACCCCUUUCCAUGGCUUGAAAAAGGUUUCCCAAGCGUCAUUCUUCCGCAUGGUAG
CUUGCAAUGCCUGCAACUCCUUUACCUAACUCCGUGUGAAAGGAAGGGAUGGGUAGGUGC
UAUCCGCGCUCGUAUCAAUUAGCAUACUACAGGAAGUUAUAUUGAAUCAUCACUUACCUU
UCCCCUAGUUCAUCAAAUUAGGCCUAUAUAAAUUAUUUCCUAUGUGAAAACUACCUACUG
AUAUGAGUUUACAGGCGACCUUGGGGAAAUAGGCAUAUAAUUUUUAAGAAAAAAGAAUCA
UUACAUCUCAAAUUCUAAUCAACUAGUCAUGAGGUAAGAACAGUGAGAAAUCACACUAUC
ACAACAGGUUAUCAAUAGGCGAGGACCAAGCAGUGUAUUCAAGACUAUAUUCAAACCACU
CACACUUGAGUACAGAAAGGCCAAACGCUUAGGAUACCCUCGUUUAACAAUAAGAUAUAU
CAACAUUCAUGCAUUAUACAUUGUCCUAAGCAUGAUAUGGAUAGUGAUUAGAGCAAUUUA
AUCUACUAGUUGACUAACCCUGUUGGUGAUCAACAGAACUCAAAAGAGCAAAAAUAACAA
AAGAAUGCUAAUAUACACAUGCCACAGAUUAAAUGUUGAUUUUAUAACCUCUUUAAGCUA
AUAUACACCUAAAAAAUGAGAAAGAGAGGGGGAGGGGAGGGGUCAAUUCCAUUAAUAUAG
AUACAAGGAAGAGCAUGUAGGUAGUCAAAAGAAUCCUUACUUAGUCCUCUAACUGAAAUU
UAGACCAAUAACUCAAGCAAGUAUAAUCAUCUUGGUGAAUUGAAUCACUAGUUUAUAUUG
CAACAUAUGAAUCCAACUAGUAAAACAAGUAACUUCAUAAUGUCAUACUUUCUGUAUUAA
CUCAACAUUGUGAGAAAAUUUUAUGCAAAAACAGGAUUUCAAUAACAGAGCUACAUUGUU
UGAUCCUAAGUACCCAUUUAAUGUCCUUCCUACUCAUUCUAGGAACACUGAUUUAAUUAU
CCAUAUUAGUCUAAAUUGCACAACAAAAGGGUCCAGCAAUAAAUUAUCUACUAAAAUCUG
UCAACAAAUACUUAAAAAUCACAGGCAAGGUCCACUGAACCAUAAAUCAGAGCAAUAAAU
UAUCUACUAAAAUCUGUCAACAAAUACUUAAAAAUCACUGACAAGGUCCACUCAACCAUA
UAAAUCAUGAAUGUUUCUAUUACUUAAAUGAUUCUCUACACCAUAUCCUACCAAAUAGGA
ACAAAAGUCAACAUAAGACACAAUAAUGCAUUUCCCAAGUAGGAUAAAUAUUUCUACGGC
AUGAAGGCAAUUAAGUCGGACUAAAUUAGAGAUUAAACAGUAUAUACUUGCUUAAAUAGA
AAAUUAGUAAAGGAAAGAUACAUGCCCAGUGUCUCAUCAUAACAAACCAUAAAUACACAU
UAUUUCAACCCUAAAAAUGAAAUAAACAGUCCAGGAUCAACUAGUAUUCACAAAAAAUAG
ACGGUACUUAAAAGAAUGGACUAUAAUAAAGGAUUUACCUUUUUAGGCCAGCAAAGCAAG
AUUAUGAAUCGAGGAAUAUUACCACUAUUUUGAACUCAAAUGUAUAACAAAAAAACCUCU
AACUCCGUGUUAGACACAAGGACUGCCCUUUUAAAGAAAUGUAAUGGGAGUUAGAGAGAA
CUCGAAUUUAACUCUAAGAUUAUAAAACGCAUAAAAUUAAAAUGUCUAUCUAAGAAUUAA
AAAAAUGAACCUAAAGCUCUAUUUUCUUAUUGUCAUUUGGACUAAAACCCUACCCACCCU
AAAAUUGUCUCCCUAAUAUUUAGUCUAUAAUAUUUGUUGCGAUUUUAUACAAACUAGUCG
CUGCCCUACUAUAAAGAGGUAAGAGCUCUAUUUAUAGAACAAAAUUUGGGCAUGACAACA
GGGGGAACACUAAUGGAAAUUUUUGAAUUCAAAACGAGUGAGGAGAGGAAAAUUACCAUU
UGAGAAGCUAGGAGAUUCUAGAAACCUCUAGCACACCAAAAUUGUACAGCAAUGGAUGAA
AAUUGCAACAUUAUGUUGAAGGAAACAUUUGUUCAUCAGCUUCACAUCUUCGAGGAACGA
GGAAAUCGAUUGCGAGGCGCGUAGGUUAAAAGGCUGCCGCUUUUGGGCGUGUGUGUGAGU
GUGUGUG
2. MFE structure-
.....(((((((..((.....))..))))))).......((((((...(((((.(((..((((.......(((.........))).....))))..)))(((((((.((.....))..)))))))...((((....((((.(((((....((((((((((((((((..................((((...........)
))).............((((((((((((((((..(((..(((...(((((......((((......)))))))))....)))..)))..((((((...........((((......)))).(((.....))).....(((((((....((((((..((((......)))))))))).....)))))))..........((
((((((....))))))))(((.((((((.....)))))))))(((............))).......(((((((((((((.....(((..((((((((((....(((((((.(((((.((.(((.(((((((((((.(((......(((((.......(((((............)))))(((((..((((.((.(((.(
((((....((((((...((((((.(((.((.....))...)))))))))...))))))((.....))((((((((((..(((....)))..(((((......(((....)))......)))))(((((((.....)))))))..((((((..........))))))............))))))))))...))))).)))
.)).))))...(((((.................(((((.............)))))..(((((((((...............((((....))))((((((..((((((.((((..((((((.((.((......((((((((((.....................))))))))))....))))...)))))).)))).)))
.)))...................(((((((((.................)))))).)))))))))(((((((....))))))).)))))))))......((((((.....))))))..........((((...............)))).........)))))..(((((((((((((.........(((((((......
......................)))))))(((..(((((((......((((..(((...((((.((((((((((...(((((..(((......((((((((.(((..((((.((((...))))...........(((((((((((.....(((.((..((((((((.((.((((...((((((((((((((....(((.(
(...)).)))...))))).)))).((((.....))))...((((......)))).........))))))))))))).)))))).)).)))....(((((((((((((((((.((.(((((((....((((((.......))))))..((((((((..........(((((.((((((((...((......))...)))))
...)))...)))))..(((((...................)))))...(((.......))).))))))))........................(((((...................))))).(((.......))).....((((((....((.......))....))))))............(((((.....)))))
......(((.......))).......(((.((((....((((((.......)))))).....)))))))))))))).)))))....))).))))))))))).)))))))))))...))))..)))))).))))).((((((.......))))))..((((((.................((((((((..........)))
)))))..((((((.........((((((..............))))))........)))))).....(((((.....)))))....((.......))....)))))).(((((...............((((....))))................((((((((((((((...))))((((.....))))..........
......))))))))))....))))).(((((.(((.((((........((((.........))))..)))).))).)))))....))))))))..)))))...)))))..)))))))))))......)))).)))..))).............)))))))))))))............((((((...........)))))
).((((..((((((((((.......))))..))))))..........)))))))))...)))))....))).)))))))))))))).)))))...)))))))))..)))........)))))))..))).)))))))))))))...))))))................(((((........)))))))))))).))))))
)))...))))).)))))))).)))....))))))))).)))).........((((((((.((((.((...)).))))))))))...))...)))))..))))))...
You can download the minimum free energy (MFE) structure in
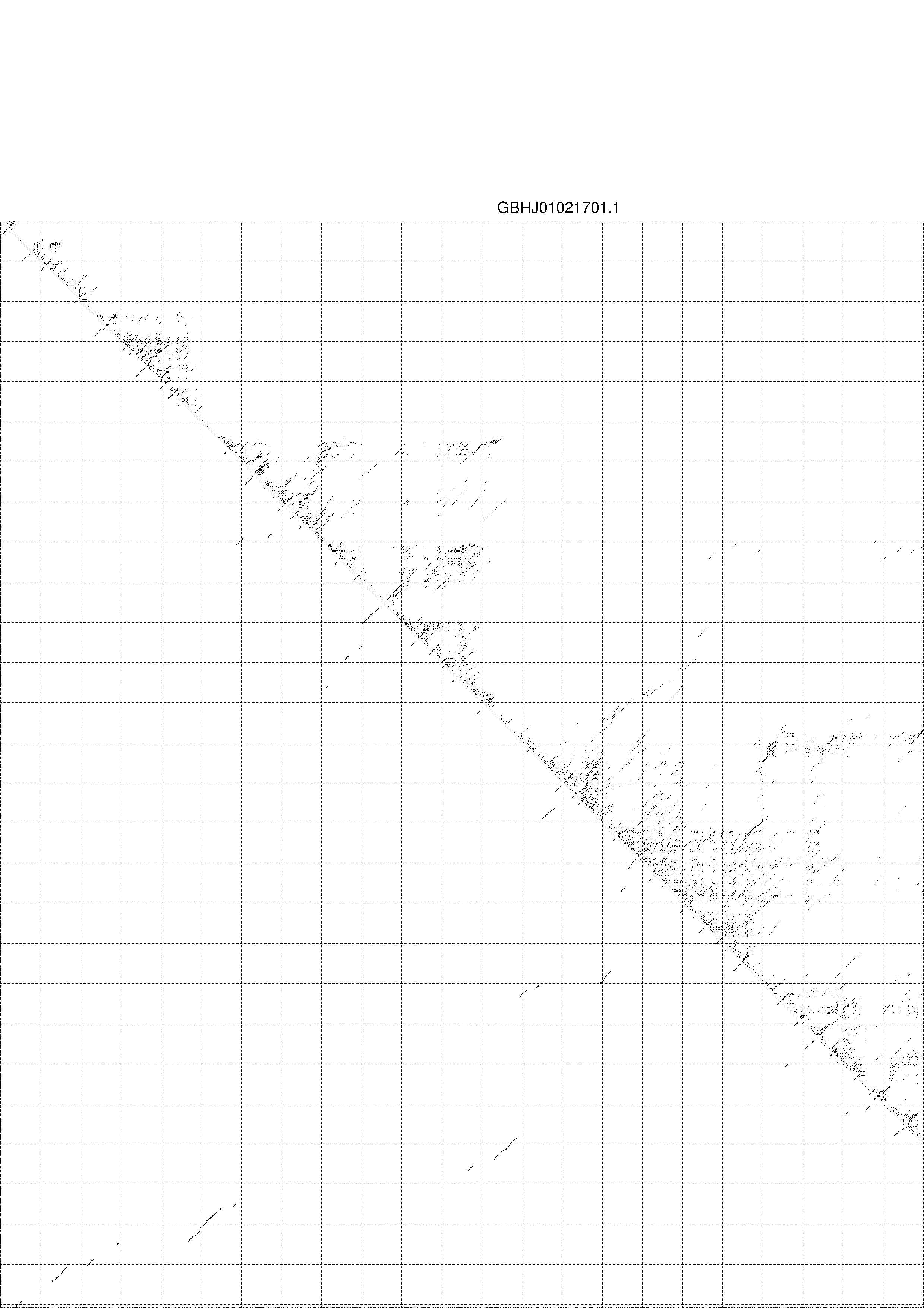
II. Results for thermodynamic ensemble prediction
1. The free energy of the thermodynamic ensemble is [-600.33] kcal/mol.
2. The frequency of mfe structure in ensemble 1.09597e-36.
3. The ensemble diversity 578.51.
4. You may look at the dot plot containing the base pair probabilities [below]
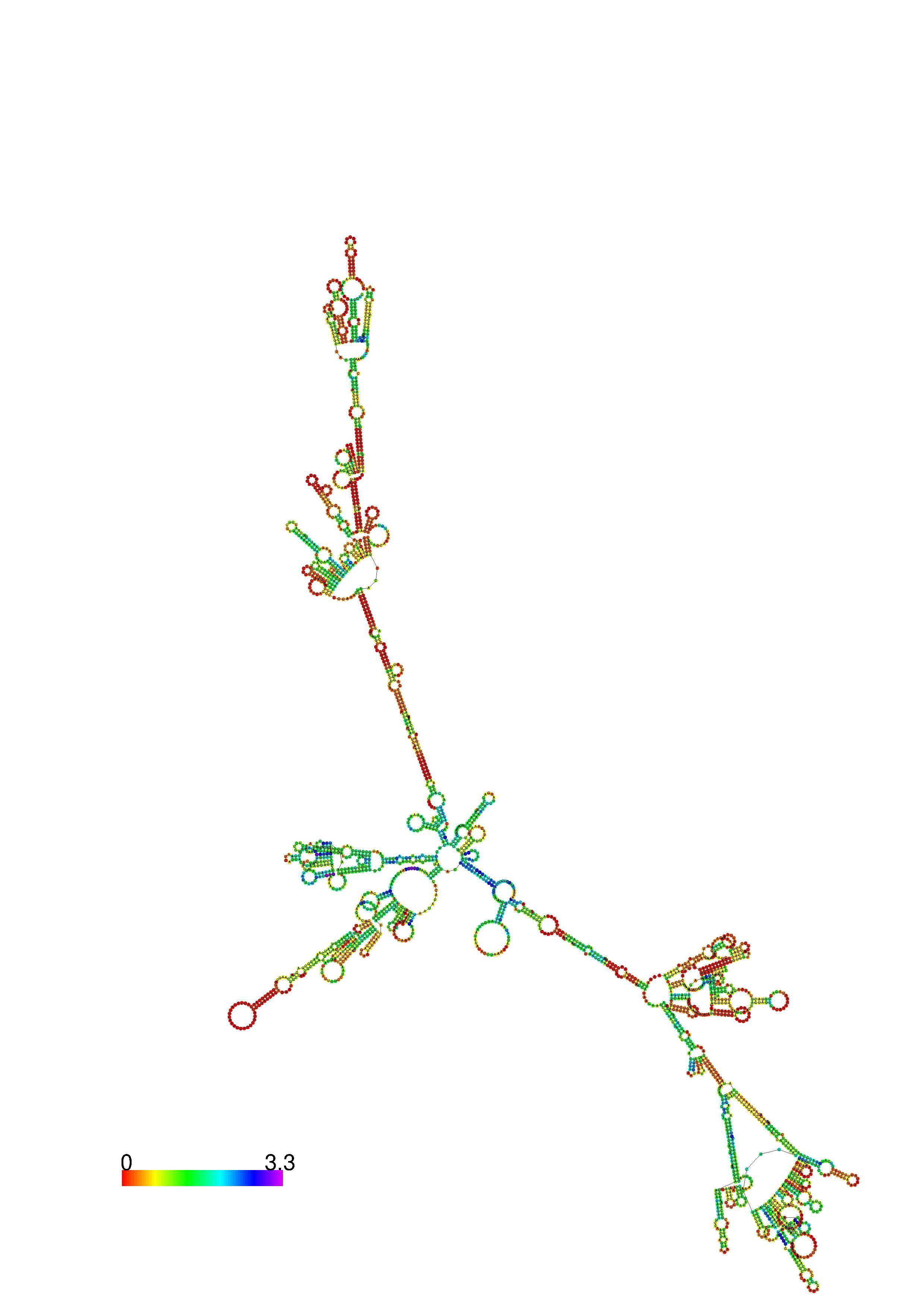
III. Graphical output
1. A note on base-pairing probabilities:--
The structure below is colored by base-pairing probabilities. For unpaired regions the color denotes the probability of being unpaired. The reliability information of RNA secondary structure
plot in the form of color annotation, Positional entropy ranging from red (low entropy, well-defined) via green to blue and
violet (high entropy, ill-defined).
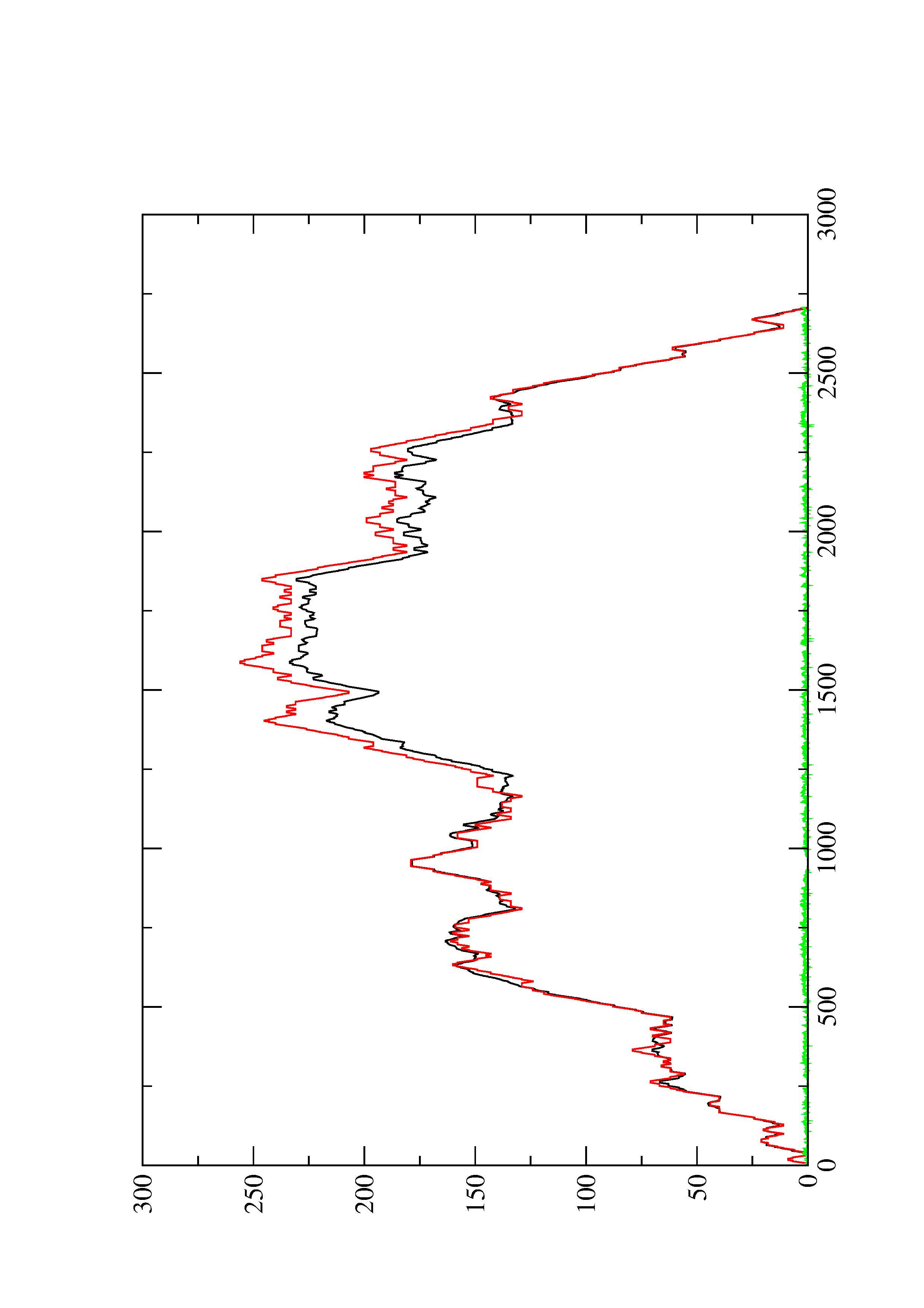
2. Here you find a mountain plot representation of the MFE structure, the thermodynamic ensemble of RNA structures, and the centroid structure. Additionally we present the positional entropy for each position. The resulting plot shows three curves, two mountain plots derived from the MFE structure (red) and the pairing probabilities (black) and a positional entropy curve (green). Well-defined regions are identified by low entropy.