lncRNA Structure Prediction
Species Name : Withania somnifera Sequence Id : GBHJ01021924.1
I. Result for minimum free energy prediction
The optimal secondary structure in dot-bracket notation with a minimum free energy of (-510.80) is given below.
1. Sequence-
AAGGGAACUCCAACCUCCACAUCGGCUCCUAUAUCUGCAAAUCAAAAGUUAUCUAAUAAU
CAUAUAAAGGGUGCAUUUAUAGAGUAUAAUCGCUCUGGACUUCCCAAAACAAAUUCCAAA
CGGAUUAGAAAAAGGCUUAGGUACCACAAUUGGCGGGACGCUCUAAGUGGUGCUCCACGC
UUGCUGAGUUUCAGGCAGUGCACUUGGGAUUUGUCUGUGUGAUAUCAAUGCACCUCAUGC
ACAUGACUAGAUGCCACAUCAACACAACAUGGCCAGUGAUACCUAUAGCUAACUUCUGUG
CAUGCUCUUUGGUGUUGCCCAUGCGUGACACACCUGCUAUUGGAGCUUCUUGAUGGCCAC
GCAUUGAGUAACCUGAUGGAGAUAUCUUCCAUUUUAUCCUUCUUAACGGGCCCAACUAUC
CCCUUUAACCAGUUGGCCACAUGCCUUGGUCAGAAUAAGAGAUUGACAUUGGUAGAGACU
GUUAUGAUGCUUUUAGAACCAAGGUCCAUAUAAACUUUCAAAAUUUCGCCGAAACUUCUA
ACUUUUAAAAAUAAUAAUAAAAAGUGAUUGCUUAAUUUUUAGAAAAUCAAUUGAAGAGUU
AUUCUUUUAAAAAAUUUAUUAAAAAAAUUAUAACUUAGGUAAAGGUUUAAUUUAAAUUCG
AAAGGCGUUAGGCACUUUCAAAAAUUUACAAAGUGUGAUUCCCGUGAAUUAAAACUUAUU
UGGCUAAUUCUAAAAACUCUUGAGUUAUUUGUAAAAAAAUUAUUAAAACGAGUUGAAAAA
UUCUUAAAAUUUAUAACGGAAAGAAAAACUCUUUUAUGAUCUAUUUUAAAGAAGAAAAUU
CUUUUAAAUUAUUUGAAAUAAAAUUGAUUUAUAAGUUAAGAAAAGGAAAGCGGUUGGAGA
AAUCAAGGAUGAUUUUUAGUUUAUGAAGGAAAAAAAGUUUGACUAAGUUGAAGAUUGAUC
AUGUCCAUAUGAAAGUGAGAGACAGUCAAAACAAAUGGAAUGCACUUACUAAAACACUCU
UUUAAGAUAAAAGAUUUUUUUUUAAAAAAUAAACCUAUAAAUAACUAAUUUUUUAAAAAU
AAAAGACAAGUUUUUGGCUUCUUUAGCACAUUCAAAAAAUAUAACUUCGGUAUUCAAUCA
AUUUUCAAAUCGUAUAUAAAGACUUCGAAACAUGAUGGCAUCCUUAAAGUUCUUAAAACA
AGGUGGUUAAUUGUUAAUUCCAUUCUUUAAACAAAAACAUCACCUAAAUAAGUAUUACAA
CUAUCACUCAGAGGUGAUUUCAACAAUAGUACACAUACGAAUGCGACUAUACUUAUAUGA
AUUUUAUCUUCACACACAAAAAAAAAACUCAAUCUUAUGACUGAUUUAAAAAGAAACAUU
AACUGUUGAUAAGUGCGUAAAUGUGGAUGAAGCUCACAUAUUUUGAGCUAUAAUUUACGG
GUGAAUUUUCAGUUUUUAUUUAGAAAUGCAUGCAGUGAAGAGAACUUUUUUUUUUUCCUG
AAAAUAUCCAAGAAACCUACAAACAUAUUCCAAGAAGAGGUAAGGUUGAAUAGAUGCACA
UUCACAUGAGUCUAUUUCCCUUCAAUUUCACGACUAACACGUAAGCUAAAAUAUGGAAAU
AAACAGAAGACUCUUAGCUUCAGUCAUUGAUAAAAUAAUAUUGUGACUUUACUUAUACUA
AAGUAGGAAAAAUGAGCAAGGACGUCCCAUGCUCAACAUUCCCAAAUGCCAUCACAUAAU
UUAUCAGAAAAAAUAAUAUCGUUGUUGCAAAAUAUGAAAUAUUUUAAACCCAAUGUAUGA
AACUUAAUCUUGAUCGAAUGAACCAAAUUUGGACAUUUUAGAAAUUCAUCCAUACAUACG
AAACAAGAUUUAUGCAACACAAUCUCAAACCAUGGAAUCGACACUUAACUCUUGAAAAAA
GAUGAAAUUGAAAAACUGAACUAUUAUUCAACAUUCAGAUACCAAAAUUCACAUAAAAGA
AAAACAAAUAAAAAAAAAAUAGAAAGAAAGAUCACCUUUUGUCGAGCAGCAAAACUGAGA
CGUGAGAGGAUAAUUUCACCACUUUGACUUCGACGUGAUUUUGUCAAAAUCGCAAGAAAA
CUCAAAUAAAUGAACUCACUGAAUUUUCAAAAUUUAAAAUAUUAGAGAAAAUUUUCUUGU
CUAAACAGUAUCUACAGAGGGUGUAUUUAUAUGGGAGAUUGGGGGUCUUCAAAUGGGAAC
UCCAUCAAUUGUGGGAUUUUGAAAAUAUCAAAAAUUCCUCAGGAAGACUUCAGCUAGGUU
AUGAUAUGAAAACGCACCAUUAUCGCACAAGAUUUGAAAAGAAUCUGGCAAAUUUAAAAG
AAUAUACUGUUGGUCGUUCGUACAAAGGCUAGAAAUUGCCUAAAAUAGCCCUCCAACGAU
CUGAUUUUUAUGGCUUGGGCUUGAAAUUGCUGGGGUAGAAGAGAGAAUAGAGGCGACGAC
UUACAGAGAAUUAGGGUUCACUUGGAUAUAUUUGCUGGAAAUGAGCGAUUGAAGAGGUCU
UGAUUAAAAUAAUAAUAUGUCGGGUCGAGUCAACUGAUUUGAGAUGGUUGGCAUAGAUUU
AGGAUAUUUGAAA
2. MFE structure-
.(((.........))).....((((.(((((.((((((..(((((((((..(((((((..........(((((((((..((((((......))))))((..((.((..(((((((((((..............((....((((((((....((((...))))....)))))))))).((((..((((...))))..))))
...)))))))))))...)).))..)))))))))))(((....)))....((((...))))..(((..(((((...((((((((...(((..((....))....)))....))))))))))))).)))..........))))))))))).)))))((((.((..(((((......(((((((.....))))))).......
.)))))))))))((((((((....((((.((((((((.....((((((((.....((((((..(((((((......)).))))).....))))))..))))))))....................(((((.....((((..(((((..........(((((((..(((((((((((((.....(((((((((((((((((
.(((((((.....................((((((((((....(((((....((((((.(((((((.....)).)))))...))))))..((((.((((((....))))))..))))..((((......))))))))))))))))))).....................(((((....)))))...............))
))))).)))))))).......((((((((((((((....))))).......))))))))).)))))))))..))))))))....(((..((.(((.(((((...(((((((....(((((((.(((((...........(((.....((((((((((((...((((...((((.((((((((.((((((((.........
.................(((((((...)))))))(((.((((((((((((.................)))))))))))).)))........)))))))))))))..))).))))..............)).))...)))))))))))).)))..........))))))))).)))...)))))))....)))))..))).
(((((((...(((((..............)))))....)))))))......((((((.....((((((....))))))........))))))...(((((..(((((((.((.(((((...(((((......................(((.((....)).)))..........((((.....))))(((((((((((..
..((((((..((((((.......))))))...))))))..((..(((((((..(((((..............(((.((((((((.....)))))))))))....................................(((((((((..((((((((.((........)).)))))))).))))..)))))(((.......)
))....)))))..)))))))..)).((((........))))((((((.((((......)))))))))).(((((......)))))((((...((((((.((.....)).))))))...)))).......(((..(((((((((...................(((((((((((((...))))))).......(((((((.
.......(((.....(((((..(((....))).))))).)))........)))))))................))))))(((((((...(((((......((((((...((((.....)))).(((.(((((.((........)).)))))..)))((((((.....(((........)))...................
.....((((((((((.....(((((..((((((..((....((((((......)))))))).)))).))))))))))))))).))......((((((((((((.((((...........(((....)))..........)))).)))....))))))))).......)))))).....)))))).....)))))))))))
)(((((((((..((((.....))))...(((.((((((((((.....)).)))))))).))))))))))))...)))))))))..)))...))))))..)))))((.((.(((((.....))))))))).....)))))..)))))..))))))))))))))...((((........))))......))..)))..))))
)...))))))).((((.(((........))).)))))))))..))))....)))))(((((((((...(((((((..((.(((.((.....(((((.......))))))).))).)).)))))))............))).)))))).))))))))..))))))))))))))..)))).)))))...))))..
You can download the minimum free energy (MFE) structure in
II. Results for thermodynamic ensemble prediction
1. The free energy of the thermodynamic ensemble is [-567.16] kcal/mol.
2. The frequency of mfe structure in ensemble 1.92016e-40.
3. The ensemble diversity 809.01.
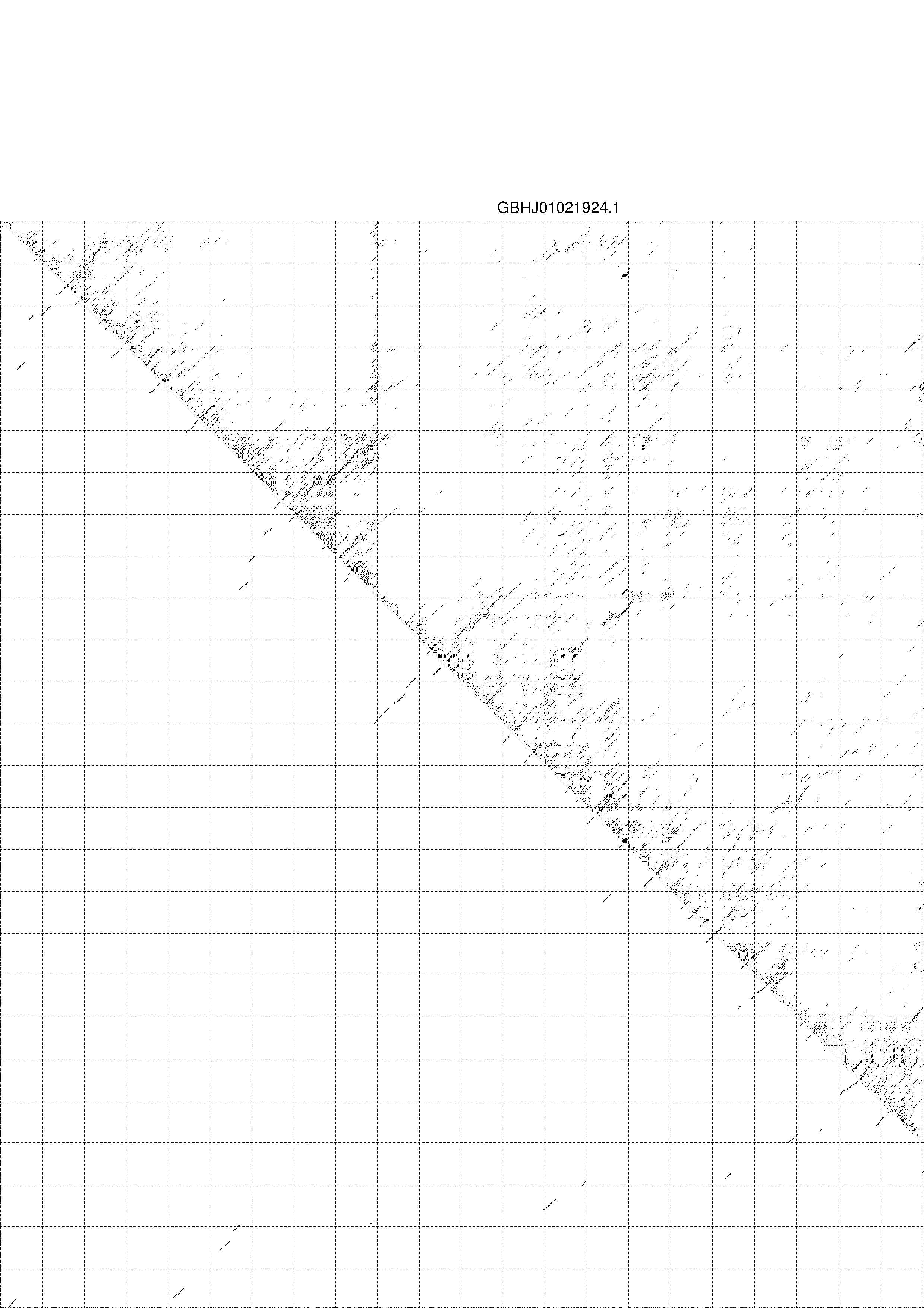
4. You may look at the dot plot containing the base pair probabilities [below]
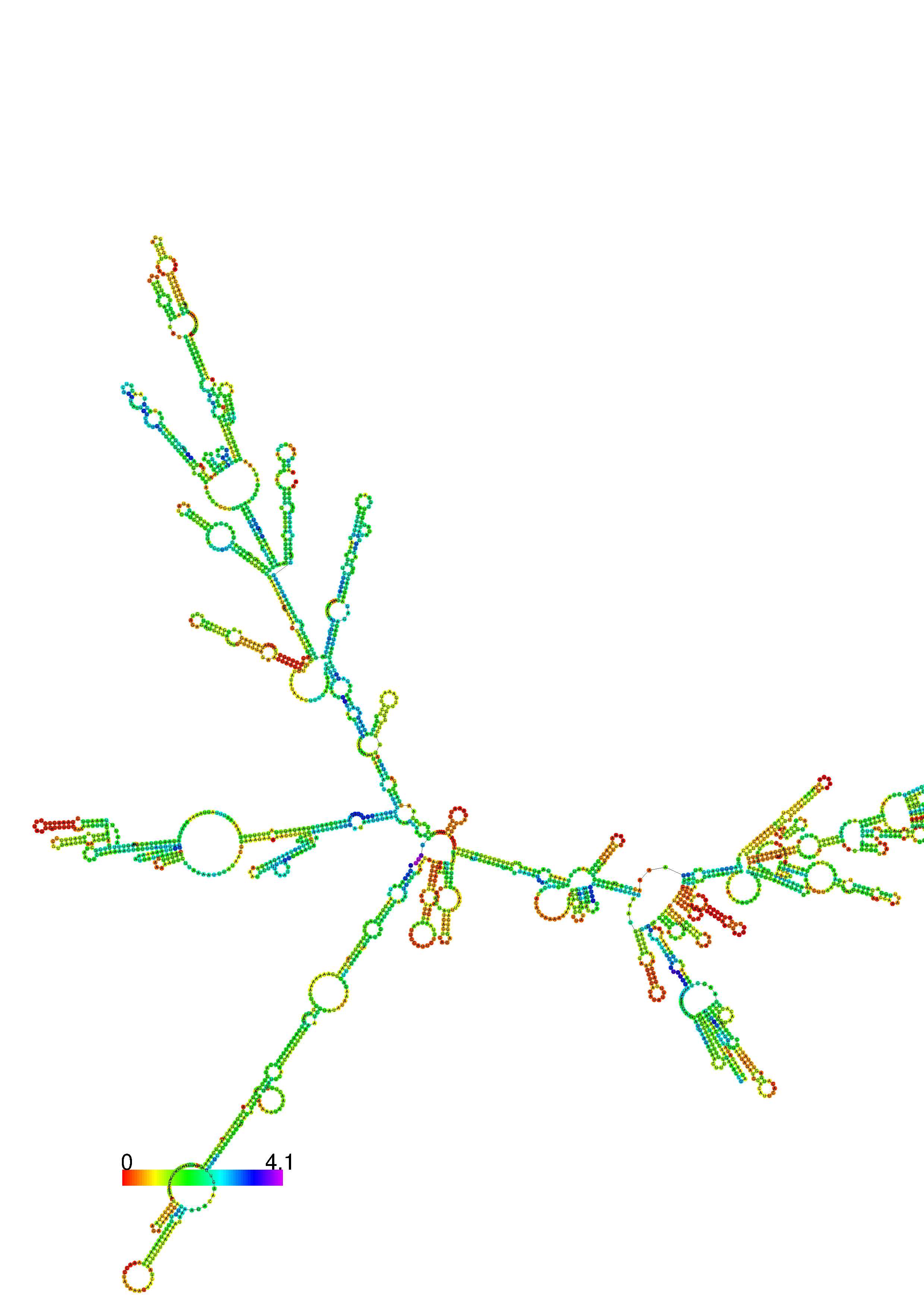
III. Graphical output
1. A note on base-pairing probabilities:--
The structure below is colored by base-pairing probabilities. For unpaired regions the color denotes the probability of being unpaired. The reliability information of RNA secondary structure
plot in the form of color annotation, Positional entropy ranging from red (low entropy, well-defined) via green to blue and
violet (high entropy, ill-defined).
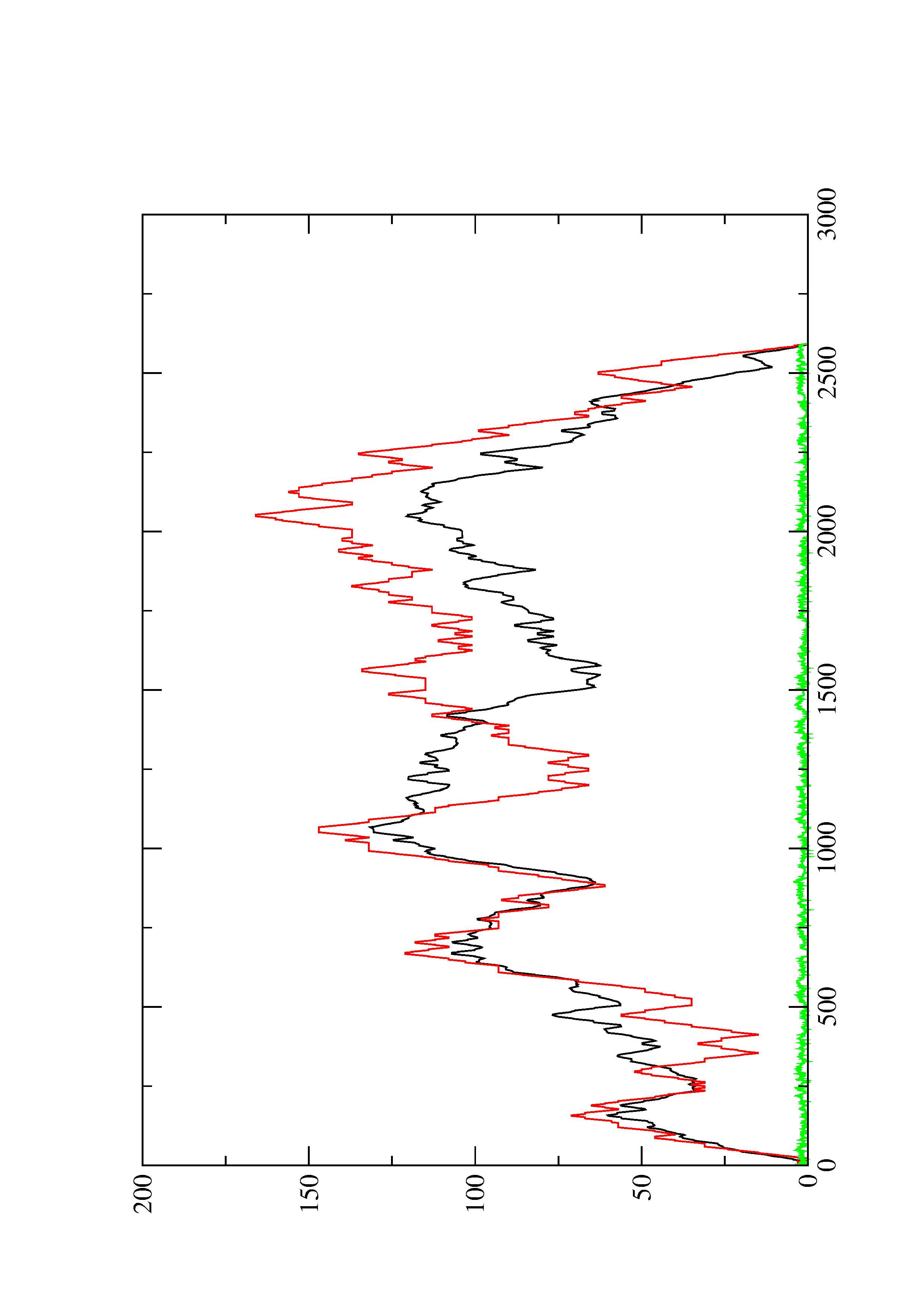
2. Here you find a mountain plot representation of the MFE structure, the thermodynamic ensemble of RNA structures, and the centroid structure. Additionally we present the positional entropy for each position. The resulting plot shows three curves, two mountain plots derived from the MFE structure (red) and the pairing probabilities (black) and a positional entropy curve (green). Well-defined regions are identified by low entropy.