lncRNA Structure Prediction
Species Name : Withania somnifera Sequence Id : GBHJ01022411.1
I. Result for minimum free energy prediction
The optimal secondary structure in dot-bracket notation with a minimum free energy of (-618.70) is given below.
1. Sequence-
CUUGGACUUGAUUAUCCUUGUCAUAUGAUAACUGGAUCCGAUACGAUUUCCUUGGGUACC
UGUCCCAUGUCUAACAAUACUAGGCUUCUAGCCUACAAUAUCGAAAUAAUAAGUAAAGAA
GAAAGUGGGCCUUAUAGCCCAUCAUCACAUCCCAUGAAUAAGAAUAAAAUAUUAUAGGAA
AUGAAGAUCGGCUUUUGGGCCCUAGAACGGUAUAGCUGUUGUUGCAAAUGGGUUUGGACC
CGCCUUUACUCUUCUCUUUGUUGAAUUAUUACUAGCCAAAAAUAAUGGGCUUGAGCCCAU
UUCUUUUGUUGUCUUAUCUUCAGUCCAUAAAGGACUUUACCUGGAGUUUGACUUGAUAAA
AUAAGAGGAAGAAAAGUCCAAUUGACUCGGAUGGAGGUGUUCAUGCAAGAAAAUGAGAUG
AAUAAUAGAUACCAACUUGUGAUGAGAAAUAAUCAUAUAAAAAUUUCAAUUAAAUAGACC
ACAAAUGUUCUAUACUUCCCGAUAACAAGUAGCAGCAGUAGUACCAUCAUACUCACAAAA
GAAAGCCAAAUUUCCUGAGAAGGAAGCAUGUAUAAAACCAGUGACCUAGCGGAAUAGCCA
CAGCAACCUAUAGGUUGUUAACUAACAAAGUCCUCCAUCAAAAUAGAUAGGUUGUUAACU
AACAUACUCCUCCAUCAAAAUAGAUGCAAACAGCUAGGAAACAGGUCAUGAGGCCCUGAA
UUUUCACACUGGUGAAACUAGAACUUAAGCAUAUAAAAGCCAAGCAAAACAGACCAAGGA
AAACAUGGCCGAAAGCUAGAUUUGUUAUGAAGAAAUGCUACCAGGAGUAUGCAAGUUGAC
UACAUGUGAAGAAGAUGCAGGUCAAGAAGACUUUACUUGAGCGGUUCAAAACCUGCUACU
AAGCUUAGAUGAUAUGCAGAGGGUGCAUAAUGAGCCAAUUUGUCUUAAAGAUUCUAAGUU
AUCUCACAUUUAUGGUGAGAACAAAAAGAACCAAGACUUCUACCUAAUACCAAACUGAUU
UUAUUCAACAAGGUCCAAAGAAGCCAAGAAUAGUUUAGAGCCUACACUAGGUGAGAAAUA
AACCAAAAAGGAUCUAGAUAACUCAAAUAUCAAAUUCUCUCAUCCUAAACCAGUCCAAAU
CCUCACAAAUGAGACACUACCAGUUGACCUUAAUUAACCCAAGUAAACAGAGAACAAAGG
CCUGGACCCUAUGCCUAAACAACCUGUACAAAAACAAGCAGAUUCAUAAUGAAUUAGCUA
UCAUCGACACUCAGCUGAUCAUACCAAAGGUAUGCAAUUAGUCAUGGUUCCAAGGUUAUU
UAAUGCAAGAAAGGCUCCAGCUCUUGACCUUUAUUUAAACAACAUGCUAAAAAGAAAAGA
AUGAAAUAAAAUCAAGGCCUUCAGCACAUAAAACAGCAACAGUCAUUUUGAAAGACCUUA
AAGACCUCAUUUUUAGGAAGUUCAACAGCACCAAAGCUAAUCACACUUCCUCUUAGCUCA
UUAAUAACACUCCAGUGUUUUACCAAUCAGCCACAAGCAGUAAGAAAAAGAAUAUUUGAG
GAGACUAUAGACAUUCUAGGCACAUUGAAUUAUGAACAAAAUUCCAACAGAACAAAUACA
AGUCUGACUUAGCCCCUUUUAGACAUACCAUUGAACUUAAAUGAUGUGACCAACACAAAG
AUUGCCAGAUUCACAUUUUUCUUGUUCAAACACCCAAGUAAAACAGCAAACAAAUUACCC
UUUUGCUGAACUAAAUGUAAAACUGCUUAAUUAGUCAAAACUUAUACACGUAGGUGUUUA
AGAAAACAAGAAAAUGAAUAACUAAUCGUAGUCUACGCAACACUAUUCCAAUCAUGCAAA
CCUCAUUAUCAGGCAUAUCUUGUCAUAGAUGAGAGAAGAAUACAGUCCCUAGCAUGCUCC
UAUGUAGGUGAACUGUUAAAUAUAAGAUGCAUGUUCUCGAUAACAUGGCAGGGUCUAUGU
CUAAAGCAAAUGAGAUAUGACACAAAUAGUCAAUUAAGUACUCAGAUAUGAACACAGAAA
CAGGUCAAAAAAUCAUCAAGGGUGAACCUUCAUUCAUUUCGUUCCAAAAAGGGCAGACAG
UGAAGUGUGUGUGGUGUUUCAGUCAUCCUACUUGGACCAUAUGAGUCUAAAAGGAGUCAC
GACAAGGUGUUCUAAUAAUCAUGCUUUACUUACUCAAACAAACAAGUAGCAGGGCUCUGU
GCCAAGCCAAAAGUAAAGAAACAUGUUGAAAACAACUACCAUAGAAAAGGAUUGUUGGUU
CUGUUCCCUGAGACUUAGGCAUGCAUGCAUAUACACCUCAAGUGGCACAUAACACAUCAA
CAAUUAGGGUCAUCCUAUCAUUAACGGAAUGCCCAAAUAGAAACAAAAAAAAAUGCAAAC
UGAAGAGGAAAACUCAAACAGCCUGAAACAUAGCAACACAUGCCAAGGACAACAGCUGAA
GUAACUAGCUAGCCAUGUUUGGCUCACUUUAUACUCAGUCUUCAAUCUAUAUUCUAAUAA
AUCAUGUUUUGACUACAACUUCAAAUCAGAAACUAGACAUGAUGAUUCACAGAAGAUGAG
AUCACGCAGUUCAUUUGAAAAGGAUAUGCUUCUAAUCUUAAACAAAGAGUCAGCUCCAUA
UAGUGCAAUGUUCUAGUGUAGUAUAGUAUAAACAGCAAACAACAUGUAGUUAUCUAACCA
UUUAAUAGUCUCAAAUCAGCAAGUAAAACAACCACCUCUUACCACGCUUCCAGUACUGAU
CCACUAUGAGAAAAUGCAAACUGAGAAAUCUUGACAGAUAACAAGUUUUAAGAGAUAAAA
AAACAUUAUUUGCCAGGUCUAACAUAUCGUUAUGAACAUAUCAGCAUUACUAACCACCUA
AGCUUCCAAUCAAGUCACAUAGCUCAGUAUUACUUAAUCACUCAUAUCAUACUACAACAG
UCAACACAAAAAAAUCAUGAUAAACAAACAAACUAGUAUUAGUAGUGAGUAAAGUAGAGG
UUUU
2. MFE structure-
...(((.(((((((((((((((....)))))..))))..))).)))..)))..((((.((...(((((...((((((((.((((((...))))))..((((((.......................((((((...((((.(((.(((..(((.((((..............)))).)))..))).))).))))...))))
)).....))))))...))))))))...)))))...))))))(((((((((...(((.(((((((.....((((((.(((((..(((((((....)))))))..)))))((((.(((((...(((((.....))))).....(((((((.(((((.......((((((((((....((((.......((((((((((((((
..(((((.....(((........((((((.((((((.((((((........))))))...........................(((((((.(((((....................((((((......)))).))....((((......)))).(((((((((((((((..........(((.(((((.((.....)).
.(((((((....))))))).........((((.(((.(((((...(((.(((.(((((((..(((((((((.((((......))))..(((((((((......(((((((...((.((...((((((....))))))......(((..((........)).)))..........)).))....))))))).....)))).
.)))))................)))))))))..)))))))..(((((.....(((((.(((((((((........(((((((((((...)))).)))..)))).........((((((.....))))))..(((((..(((((.(((((...(((......((((((.......)))))).......((((((.((((((
((((((........((((..((((((.....(((........)))..)))))).)))).........))))).)))............(((((..(((.................)))...)))))...............((((....)))).......(((((((...................((((.......(((
(.(((...))).)))).......)))).........(((.((((((...)))))).)))...........)))))))..((((((...)))))).....)))).))))))..)))...))))).)))))...)))))....))))))))).))))).....)))))..........((((((((.........((((.((
((((.......................))).))).)))).......))))))))(((((((.....(((......)))......)))))))..............(((((....)))))..))).))).((.....))................)))))))))))).........))))))))...(((((........)
))))....((((((.......((((((((.....((((((((((......((((((......(((((((......(((...)))......)))))))......))))))....((((((((((((.((.........(((((((...((((.(((((......(((((((((........((((.((((....)))).))
))......(((..((((........))))..))).......((((((....(((((.....((((((...((((((((((((((((......(((((....((((((((((.((((....)))))))).)).)))).................)))))......))))))))))..))))))......)))))).)))))
....)))))).))))))))).(((....)))..........((((.......(((((...))))))))).....))))).))))..)))))))((.(((......))).)))).))))).........))))))).........))))))))))))))).)))....))))))......)))))))).))).))))....
........((..(((.....)))..))...))))).)).)))))((((....))))..((........))...)))))))))))).(((((...))))))))...)))))..)))))))..(((........)))...........((((..(((..........)))..)))).......................(((
((..(((.....)))......(((....(((........)))...)))......(((((.(((...((..(((((....)))))..))...)))))))).....((((...((((.....((((((((((((........))))...........)))))))).......))))....))))....))))))))))))..
.))))...)))))..)))))......))))))))))))....(((...((((.((((((.(((((((...(((.(((.......))).)))..........................(((..((((............))))...)))...((((((((....(((((....(((((......(((...((((.((((((
(...(((((..........))))).))))))).)))))))..((((....)))).........))))).............((((......))))..))))).))))))))........))).)))).)))))).)))).....)))...........))))))))).....))))))))))))).)))...))))))))
)...
You can download the minimum free energy (MFE) structure in
II. Results for thermodynamic ensemble prediction
1. The free energy of the thermodynamic ensemble is [-671.12] kcal/mol.
2. The frequency of mfe structure in ensemble 1.14718e-37.
3. The ensemble diversity 869.81.
4. You may look at the dot plot containing the base pair probabilities [below]

III. Graphical output
1. A note on base-pairing probabilities:--
The structure below is colored by base-pairing probabilities. For unpaired regions the color denotes the probability of being unpaired. The reliability information of RNA secondary structure
plot in the form of color annotation, Positional entropy ranging from red (low entropy, well-defined) via green to blue and
violet (high entropy, ill-defined).
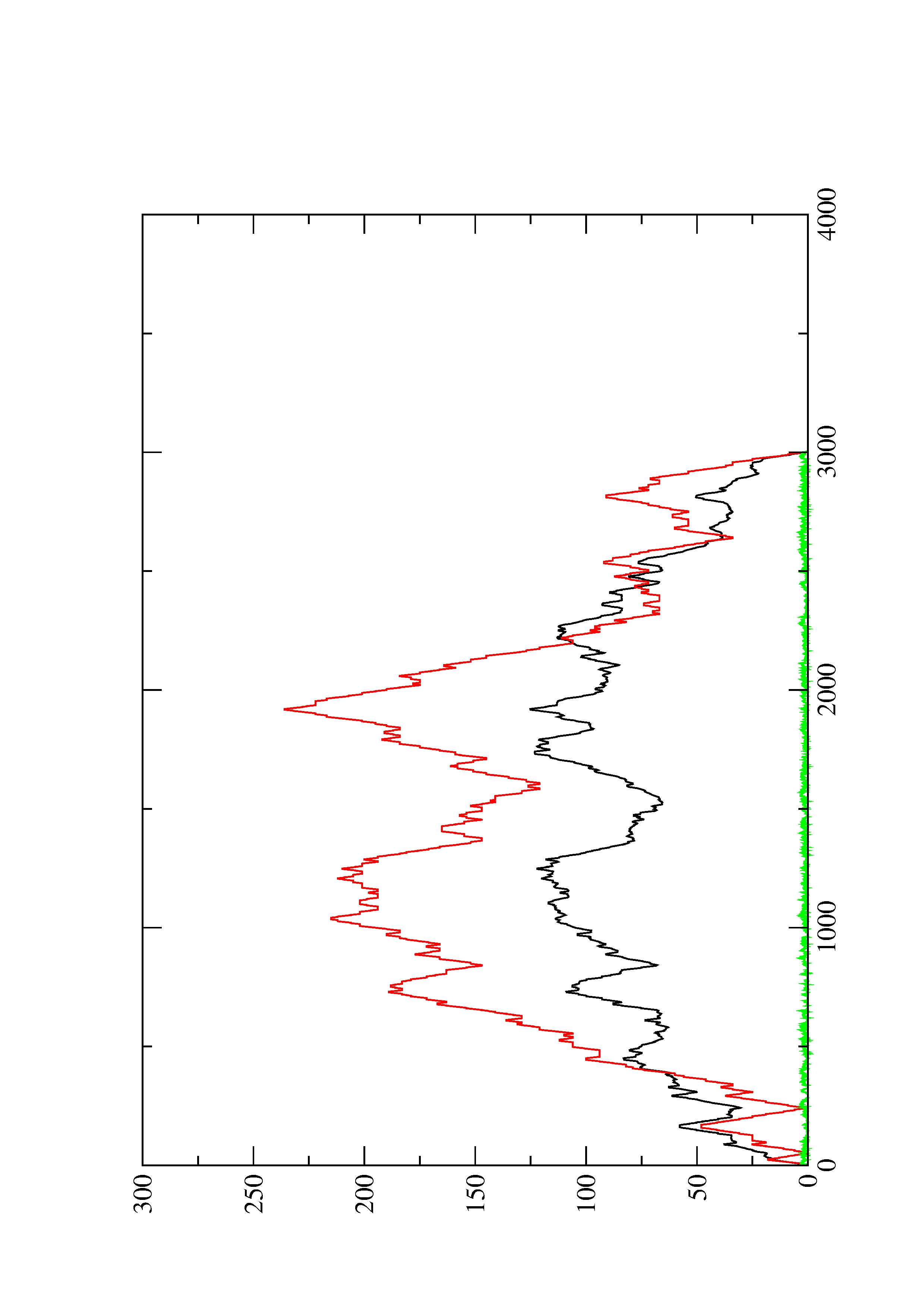
2. Here you find a mountain plot representation of the MFE structure, the thermodynamic ensemble of RNA structures, and the centroid structure. Additionally we present the positional entropy for each position. The resulting plot shows three curves, two mountain plots derived from the MFE structure (red) and the pairing probabilities (black) and a positional entropy curve (green). Well-defined regions are identified by low entropy.