lncRNA Structure Prediction
Species Name : Withania somnifera Sequence Id : GBHJ01022732.1
I. Result for minimum free energy prediction
The optimal secondary structure in dot-bracket notation with a minimum free energy of (-371.20) is given below.
1. Sequence-
UAAGGCUAAUAUAAACACUCUAUAAACUGACAAGUAAUAGGAAGGUUACACAGGACCAAU
UUAAGCUUUUAUUUGACAGUUAAAUGACUAGCAAAGCAGAAAGCAGGAAAACAAAAGGAA
GUAACAUGUCAUUUUGAACAUGUUUAACAAAUAGUAUGCAAGAGAAUUCAAAAGUAGUCC
AGGCACUCAAAAGUAAGACAAUAAGUGCAAUUGAAAUCUAAUCUAUCAGACAUUGCAUGU
UGAAAAAAGAGCAGGAACUUAAUGAGAAUCAUUAGUCGCGUUUGAAUAGGACUGGCAUAA
AAGAGCAGGCAGAACUAGAAGCAAAGAAGUCAUGCAUUUUUUAGACAUGGCAUGUAGCUA
AACAAAACCGAAACAUGUAUACAUGGAAGGCUAACAUAUCAUUUAAAGUGUGCUCAAAUA
CAUAACAACAGGUGGGACUAGCUUAUCCAUUAUCACAUGCUUAUAUUCCUUUAAACAAAA
CUAUAACCUAAUGAACAAAGUCUAACUCAGCUAUCACACUUGACUAAAUCUUGCAUAUUA
AGUCAGUUAAAGAGGAGCACAAGUAGGUUUAUUAUACAAUUUUAAGAGAAGGCAAGUAGA
CAAAAUGGCAGUUUCAAUAGAGGAACUUACUGAACAGGUCACACUAAGACUUAAAGACAA
AACAAUCAAGUAUAGGACUACUACUGGUUCUCAACAAUUUAGCAGACAACAAGGAGGUCA
UGCAUGAUCAAGUAAGCUUUGAUGACAGAGUCAAACAAUCAACAGAUACAAUUAUACACA
UGCACAAUACUUAAGAGAGAAAUAACAUGCUAAAUGCAAGCAAUCAAGCAAUCAUGAGAA
GAACCGGGAGAAGAACAGAUCUUAGUCACAAUAAUACGGAAUUAACAUUCAAUUGUAACU
UCAUUUCAGGCAAGCUUAUGUCAUUUAGACCAUCAUGUCAAGAAAAAACAAUUAGGCCAC
AUUAAGACCAAUGGAAACAAAGAAGGCUUUAGUCACAGCAUUCAAACCUAAUACAACACA
CUAUUUAACUUCUAUCUGACCAUUUUUAGGAAAUCAAACCAUCAUGUCAAGCUGGGAAGU
CAGAUUGGUGAUCAUACAAAAGAUAUUACCUAUUUUAGUUAUUUAACUUAGCCAUGGUGA
GCAUGCAACCGGUUAUUGGUAAAGGAAAACAUGUAGGCAUGUCAUAGUUAUAAAACAGUG
CACAUUUGGCAGACAUGUCAUAAGACCUAAGCACUUAGGUCACAUGGAGGCAUGGUAAAU
CACAUUUGUAGGCAUUCAAAGCAAGUUAAGUGUUCUUAACAGGAAAAGUGCACUUCUCUA
UCAGUUUUUCAGAAAGGGCAUGUAUAGAAGAGUUUUAAAUGAUAUGUUAUCCUGAGGGAU
UAACAAACUGUAAUAAUUACAACUCUAAUCAUCCCAUACCCACCAUUAUACAAGUAAGUC
UCAAGCUAGUAAACUCAGUCUGCACAGGAUUUUACUAUUAAAUUUAAAGGUCCGGCAAAA
UAACACUCCAUACGGAACUUAUCAGCAAAAAUAACAGGGAUUUAGAAAGCAUCUUACCUU
UUUAAGGGAAGCAAAUCGGGACUACAAACUGAGGAAGGAUACUACUGUUUUUUCAACAUU
UAUUAAAAACACUAAACCUCUCAAAUCUCCAGUUCCCUUAUAGACACAAAGAAAUCCCCU
AUAGAGAAAUGUCGUAGGAACUAGAAAGCACUCUGAUUUUACCUCAUCGGAUUAUAAAAA
CGUUUGAAACUUAGAAUGAUCAUCUAAGAACUCACAAUCGGAACCUAAAGCUCCACUUUU
UGUUAGUUGGAUUAAAAAUGUAACUGAAUUAAGAUUCUUCCUUAGUUCUUAGCCUAUUGU
AGCUUCUCACCUCUAGUCUAUACUACCAGCUUCUCUUAGCUGCUGCUCGUCUCCUCAUCC
AUACCAAAAAGUAAAAGGGAAAAAAG
2. MFE structure-
........................(((((.(((((((.((...((((......))))........)).))))))).)))))....(((.((((.((((((((((.((...............(((((((((...)).))))))).......((((((((.((((............((((..(((((.............
...)))))...((((((((..............(((..(((.......((((..((..(((((((...)))))))))..))))......)))..))).......((((((.(((((((.(((......((((((...........)))))).....))).............((.((((((.((((.(((.(((((((((
(((((((((((......)))).........((((((.....)))))).(((((.(((((((((((((((((((..((.....((((((((((((((.........(((..((.....(((.((((.(((.......))).)))))))...))..))).........))))))))............(((((..((((((.
..(((((((((((((......))))))..(((((.(((((.......)))))................((((......)))).((((((((.....(((((((((((.........))).((((((.........((((((....))))))......(((....)))...........)))))).....(((....))).
.......))))))))....((......))..........))))))))((((........))))(((.(((((...................))))).)))....)))))..........))))))).(((......)))...........((((((...((((......))))........(((.(((........))).
)))...))))))........))))))..)))))(((((((..(((((((.....((........)).....)))))))))))))).(((((((........))..))))).......((((.......))))((((.....)))).....)))))))).)))))))....)))))))))))).)))))........((((
(((...(((((....)))))...(((((((....))))))).((((....)))).................((.......))..((((((....)))))).......)))))))(((((((((((((((....)))))).)).)))).)))..)))))))))))))))).)))..(((((........(((((...))))
).........))))).......)))))))))).))..........)))))....)).))))))...))))))))............(((.((((..................)))).))).................))))((((((((........))))))))(((((((.(((.(((.(((..((((((((((((((
.....(((((((............))))))).........((((((.((.((((((..(((.((((.......................))))))))))))).))......((((((........))))))..........))))))..((((((.......))))))...........(((.........))).(((..
..(((((((.(((...))).)))))))...)))))))))))))))))..)))))))))...)))))))..)))).))..)))))))).)))).)))..)))..)))).)))((((......(((......)))...))))......
You can download the minimum free energy (MFE) structure in
II. Results for thermodynamic ensemble prediction
1. The free energy of the thermodynamic ensemble is [-408.96] kcal/mol.
2. The frequency of mfe structure in ensemble 2.4548e-27.
3. The ensemble diversity 352.69.
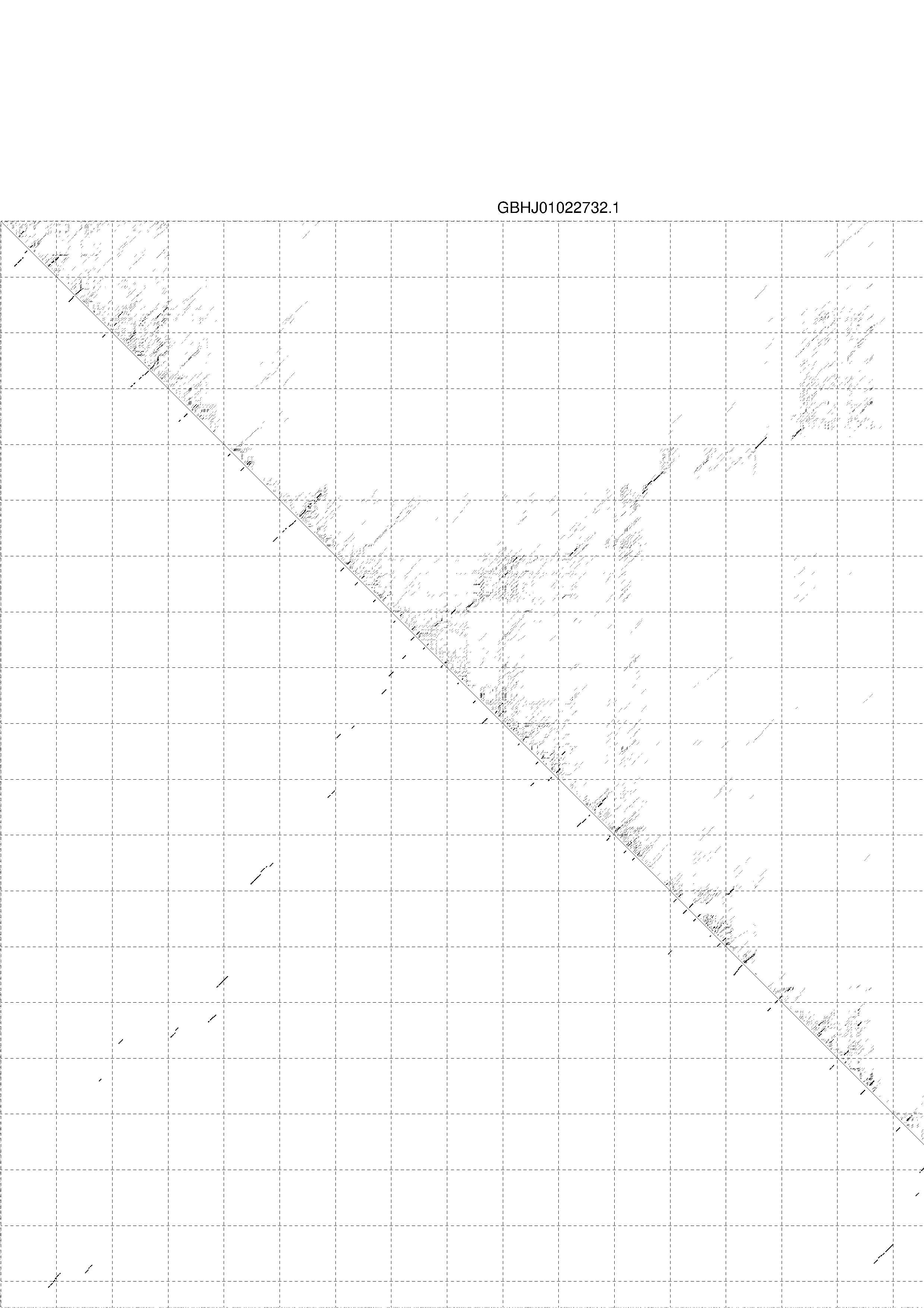
4. You may look at the dot plot containing the base pair probabilities [below]
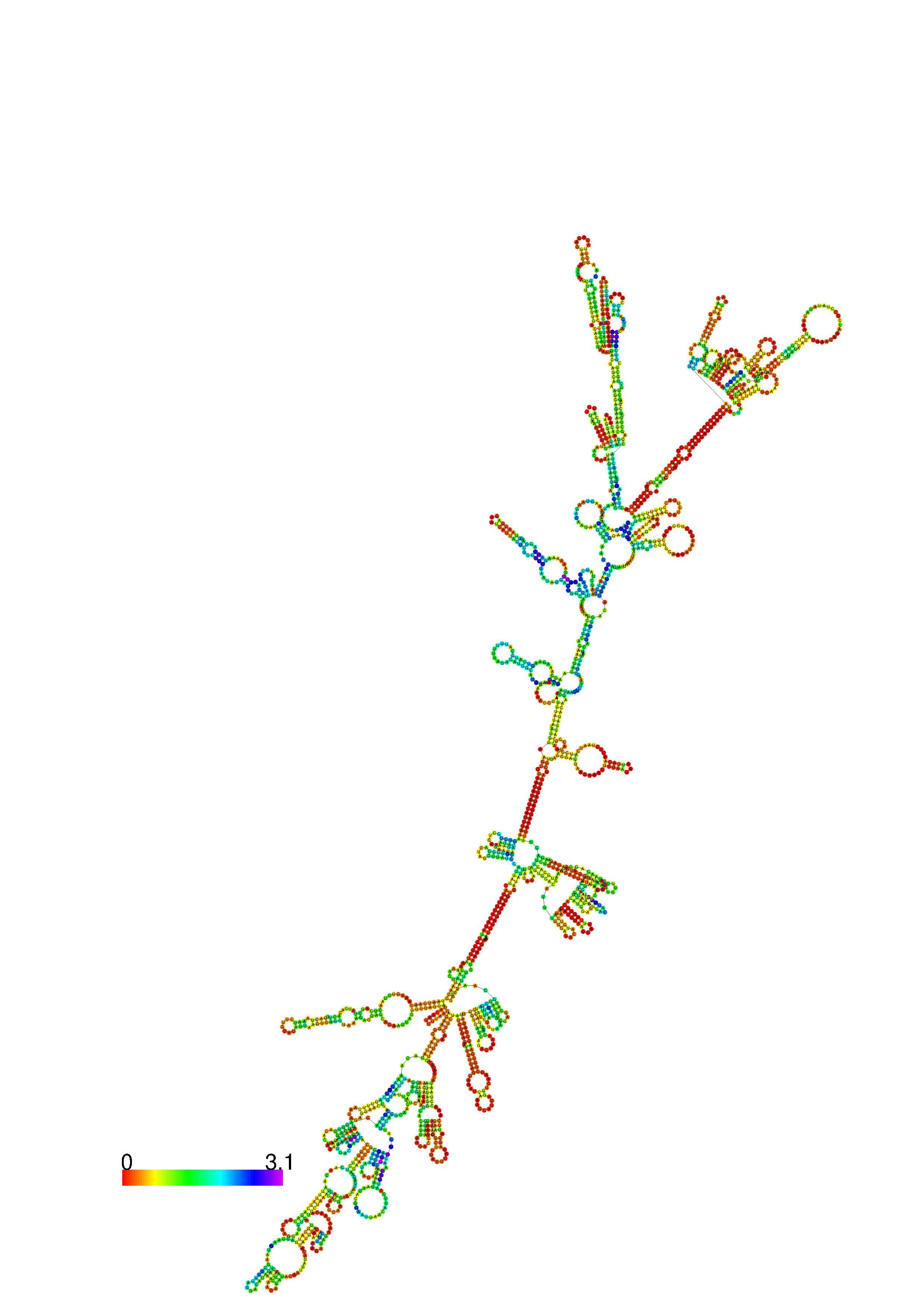
III. Graphical output
1. A note on base-pairing probabilities:--
The structure below is colored by base-pairing probabilities. For unpaired regions the color denotes the probability of being unpaired. The reliability information of RNA secondary structure
plot in the form of color annotation, Positional entropy ranging from red (low entropy, well-defined) via green to blue and
violet (high entropy, ill-defined).
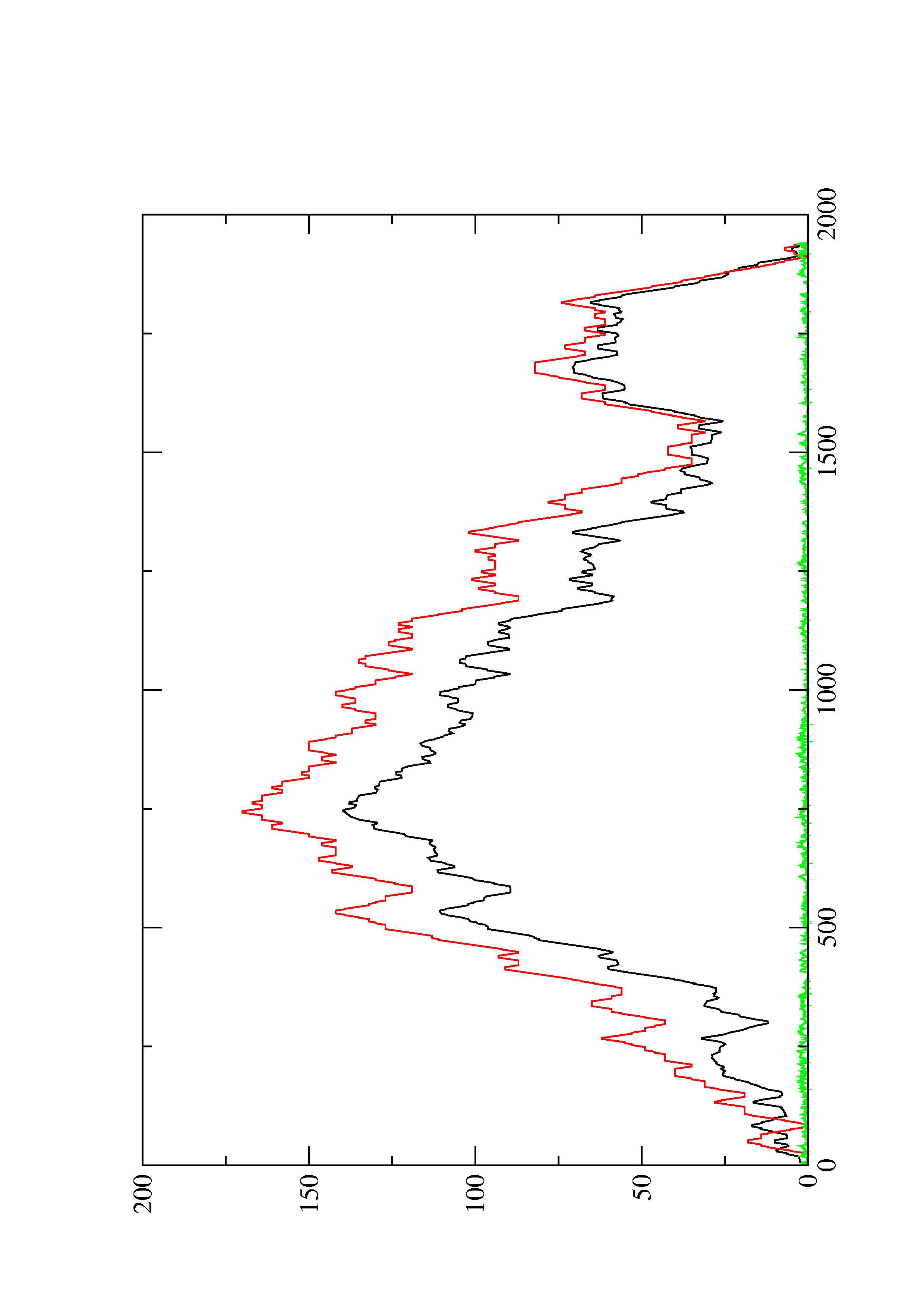
2. Here you find a mountain plot representation of the MFE structure, the thermodynamic ensemble of RNA structures, and the centroid structure. Additionally we present the positional entropy for each position. The resulting plot shows three curves, two mountain plots derived from the MFE structure (red) and the pairing probabilities (black) and a positional entropy curve (green). Well-defined regions are identified by low entropy.