lncRNA Structure Prediction
Species Name : Withania somnifera Sequence Id : GBHJ01023071.1
I. Result for minimum free energy prediction
The optimal secondary structure in dot-bracket notation with a minimum free energy of (-540.80) is given below.
1. Sequence-
CCCGACAUUGCCCUUCAAUUAUUCAUACCCAAAAAGGAAAACAAAUCCCCCACUCAACCU
CGUUAAUAAAACCCAACCCAACCCAUAUGUUUCCUACAACCUAGUUUCCAAUACCACUUA
UUCUAAAAAGAAAUCACAACAUCUCACAUGCAUCCCAGAAAACAUAAUGACUACAGCAUA
UUAACAUAAUUAACACAGAAAUAACCAUAAUAAAUAAGACACAUUCUCUAGAUUACGCCC
AAAUCAUAAACUUAAAGAAAGAACAACAUUUCCAAGAAAGGAGUCUAACUACGGAACUAA
AAACUCUUCAAUCUUUCAGGCAAAAGGGGGAAAAUUGUCUAAAGUCAUUGUUACUGAGGG
AGUCUAACUAUGCAACUAUGUAGGUAAAGAAUAUGGCAUCUUAAACUCUCUGAUGAUCAG
GUAAUUUAUAUUGGUACCACAACCCCCUAAAAACGUGAUAGCUAUAUGAACUUACAAGGC
CAAUUAUUUCAUCUAUAAGGUUCUUAUGUUAAGAAAGAGAAUCUCAGUUUAAGAAAUUGU
UAUUAAUCAAAGGUCGGAUAACUAUGAGAACAGAAAAAGGUCGAAGAUUUUGUUAUUACA
AUUCUUAGGUCGAGAAAAAAAGAACGAGAGACUCUUAAUGCAAGGCAUUAUUUCAUGAUU
AAUAUACAAAGUUUAACGCCAGACGUGCAGACAUCUCAAGCUUCCCCCGUUUCACCAGAU
AAAAAGGCCUUAUUUUCUCCUUGGGGGGCUUUGGGGGUGGGGUGGGUUUCAGCCUAACAG
GAAUAACGUCAGUCAAUCAAGUAACUCUCAAAUCCCAAUUACAAACUAGCUAGGGUUGAC
GGGGUAUGAUCUUCUUUUUCUAACGUUUGAAAAAUCCGUGUGGAACCAACUGCAGCCUUC
UAUACUCAGGGAUGAUGCAGCCCCUCUCCCUACCCCCCUCUACCUUUCACCACUUAAAUG
UUGGACUUAGUUCACUAGGCACAGAGCUUGAACUUGUACCAUGGACACAAGUUCCUCAGU
CUUCCAAUUGAGCUGACCCCUGGCAGCUGACGUACAGUAUGAAUCUUCUAUGUUCACUCU
AUUCCAUCCAGUCUAUUACAUUGUAAUAAGAACCACUCGCCUUUAAAGUUACAAAAUCAA
UUGUUUGUCACUACAAAAGUGUACAACAACAUGAUCAAAUUCAAUAUUCGUCCUACCUAG
CUCUUAAAUCAAACAGUGGUCCAACAAGUAAUCAAACAAUUCAGUAAGCGCCAUCAAUUC
AACAUACAAAUAAGGAAAAACACAGGAAAUCAAAAAAUAAGAAAAAGGAAGAAAAGAUCA
AAGGCAUUUCAAGCUUCCUGCUGGGCAAUAAGGCCUAUUCCUUUAUGUUUCUUUAUGUGG
GAGGGGGUAAUUACAGGGCAUAAAGUCAGGCAGUCAAUUAAGCCUCAAAUCUCAAUACCG
AACUAGUUAGGGAUGACGGUAUGUUCUACAUUCACUCCACCCCAUUCCUUCCCAUUACAA
UCUAUAAAAAUAAUUUGCCUUUGAAGCAUACAAAAUCAAUGCCUCGUAACUACUAAAGUG
UAUCACCAAAUUCAAAAUUUGUCCUACAUGGUUCUUAUUAUUUGUAUGGUCUUUUACGCU
UCUCAAUUAUUUACUAUGUCUUAUUUGAUAUGCCUUGUUCUACCUCUACAUUGUCUUUUU
CAAAUGGUCCUACUAUGUGUUUGUUGCUUGAGCCGGGGAUCUUUCAAAAACAGCAUCGAU
AAUUCCACGAGGUAGGAGUAAGGUCUGCAUACAUUCUACCCUCCCUCCCCAGACGAUCCC
ACAUGUCGGAAUACACUAGGUAUGUUAUUGUUGUUGUUGUCGUCCUACAUGGUUCUUAAA
AUCAACCUUGGUCGACCCUUUUAGAGUUACAAAAUCAAUUGCGUGUCGCUACAGAAGUGU
ACACCAAUAUGAUCAAAUCCAAAAUCUCCCCUACAUAGUUCAUAAAUCCUAUGUUGAGGG
UCUACUGGAAACAAGGUUUGCAUACACACUAACCCCAGACCUCAAACCCCAGACCUCAAU
UGUGUGAUUACAUUAGGAAUGUUGUGGUUGUUGUCCUACAUGGUUCUUAAAAUCAAGCCG
UGGUCGACCCUUUUAGAGUUACAAAAUCAAUCACUUGUCGCUACAAAAGUGCACACCAAU
AUGAUCAAAUCCAAGAUCUCCCCUACAUGGUUCUUAAAUCCUCAACGCGAGGGUCUAUCG
AAAACAAACUCUCUUAACUCUAAGGUAGAUGUAUAGCCUGCAUACACAUUAUCCGCCCUG
GAUCCUACUUUGUGAUUGAUUACACACUAGGUAUGUUGUUGUAUGGUUCUUAAAUCCUAU
GUUGCUCGUACUCCUAAAAUGCCAAUGGAUGUGUGUCCAAAUUCCUCAAAGGUAGUGUCU
UUUUAAAAAAUCCUACACAAGUGCGCCAACAAAGGCAAAGAGUCCGUGCAACUUAGCAGU
UAACUCAAUCAGUAGUCAAACAAAUAAUAAAAAAAAUAGUAAAAUCACAUCACAUUAACA
AAAAAAUCAACGCAAUAAUAGGUAAAAAUUUUACCCUCUUGAUCCAUACAUAGCUCUAUG
AAUUUCGUUGAAGAAUUCAAUCGAUUUGAAGCUUCGCUUGGGAACCCUAGAGAAAGGAAG
AGGAGAAAACCCUA
2. MFE structure-
.....((..(((((((((..........((.....)).......((((........((((.((((((((.............((((((.(((((((((...((((...............((((....))))................((((((((.......(((((((((......(((((.....(((........)
)).....)))))....(((((..((((((.......(((.................((((........)))).((((.((((((..................))))))...))))...))).....))))))...)))))..)))))))))......)))).........)))))))))))))).)))..))))))..((
(((((((....(((((((..(((((((((((.(((..(((..............)))...)))))))))).))))..)).........)))))....((((((((...........)))))))).))))))))).....))))))))...)))).))))......((((...(((.((((((.((((..((((....)))
).)))).(((.((((....(((...((((...(((..(((...(((.(((.......................))).)))...)))..)))..))))...)))......))))))).........)))))).))))))).)))))))))..))((((((((((((((....)))))...............(((((((.(
((..........................)))..)))))))(((((...(((.((((((((........)))))....((((((((............))))))))....))).)))...)))))...)))))))))(((((.((((((.((((......)).)).....((((.((((((...((((((((((((((...
......))))))))((((((......))))))..((.(((((((.((((.....)))).(((((.......)))))...............((((((((...))))).)))((((((....((((((((((.........(((((.((((((.....)))).)))))))..(((......)))..............)))
)).)))))......))))))....(((((((.((((((....(((.((.(((((...........((((...(((...((((.(((..(((((...((((((....((((((((((((((.(((((((...(((.....)))((((......)))).......((((((((.....(((((..(((((......((((((
((..(((((((..........))))...((((((((.........))).))))))))..))))))))......)))))..)))))............(((..(((....)))..))).....)))))))).......))))))).((((..(((...(((((....((((((...))))))...))))))))..))))..
......)))))))))...)))))...............)))))))))))...)))))))...))).....)))).........))))).)))))...))))))))))))).))))))).)).......(((((((.(((((((.((..((((.(((.(((.(((........))).)))))).)))).((.((((.....
...)))))).........))...))))))).)))))))((((.((.....((((.........))))..)).)))).....((((((((........((((.((((((((.....)))).))))))))..((((((................((((((..........)))))).((((((...((((.....(((((((
...............)))))))......))))((((..(((((((((((........((((((((((((...(((...(((((((..........)))))))...)))...((((((((((..............(((((((.....)))).))).......((((........))))...((.....))(((.((.(((
(((.....)))))).))..)))............))))))))))((..(((((.....)))))..))...((((.....)))).....(((((..((((((((.....((...((((((((((((((((((....(((.(((((..((((((.............(((((....)))))....(((...)))..((((..
..............)))).)))))).))))).)))..))))).)))))))))..)))).....))......)))))))))))))..................)))))))..)))))........)))).)))))))...)))).........))))))))))))......))))))))......((((((((...)))))
.)))....)))))).))))))))))......)))))).)))))...........
You can download the minimum free energy (MFE) structure in
II. Results for thermodynamic ensemble prediction
1. The free energy of the thermodynamic ensemble is [-588.65] kcal/mol.
2. The frequency of mfe structure in ensemble 1.91838e-34.
3. The ensemble diversity 489.78.
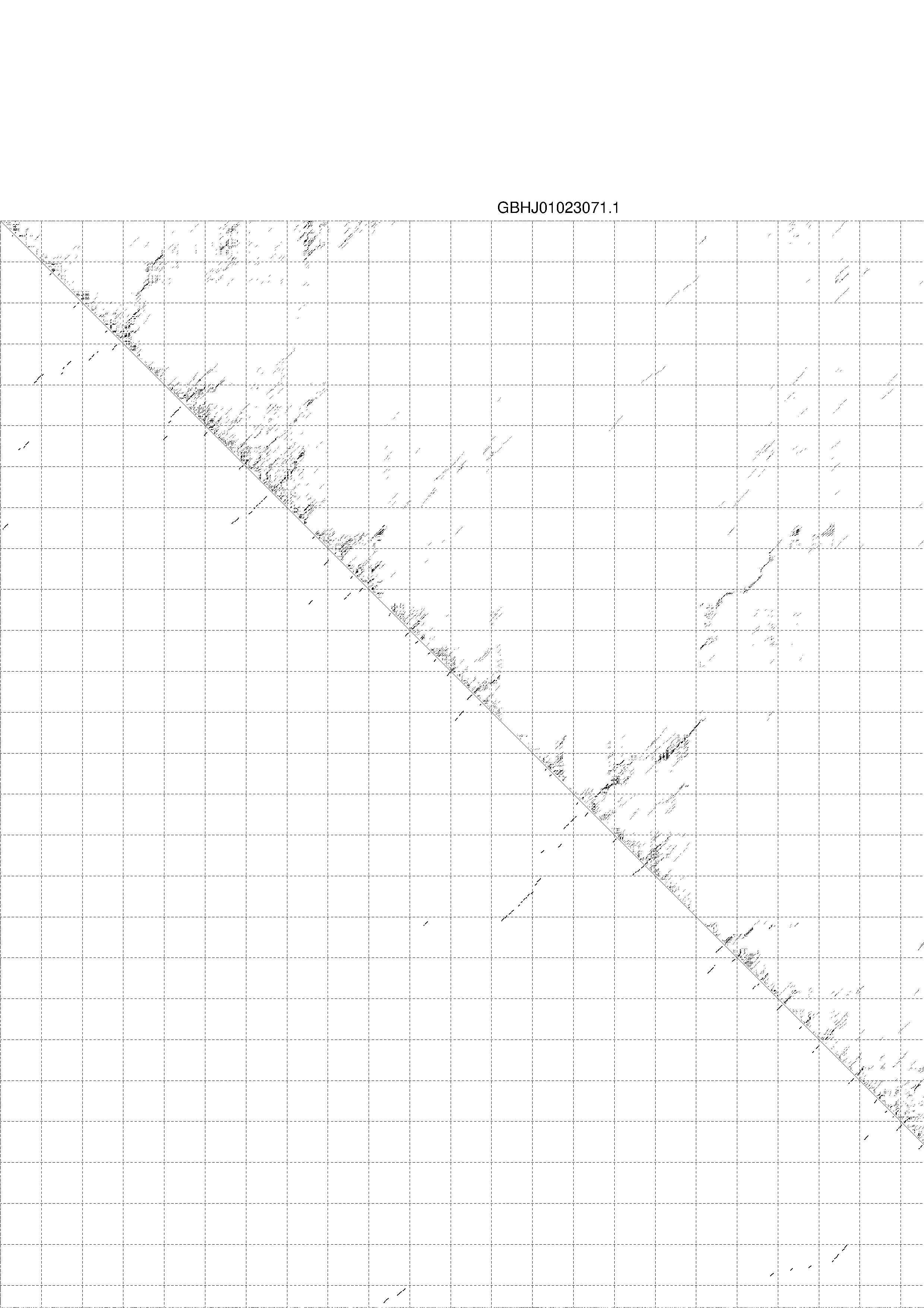
4. You may look at the dot plot containing the base pair probabilities [below]
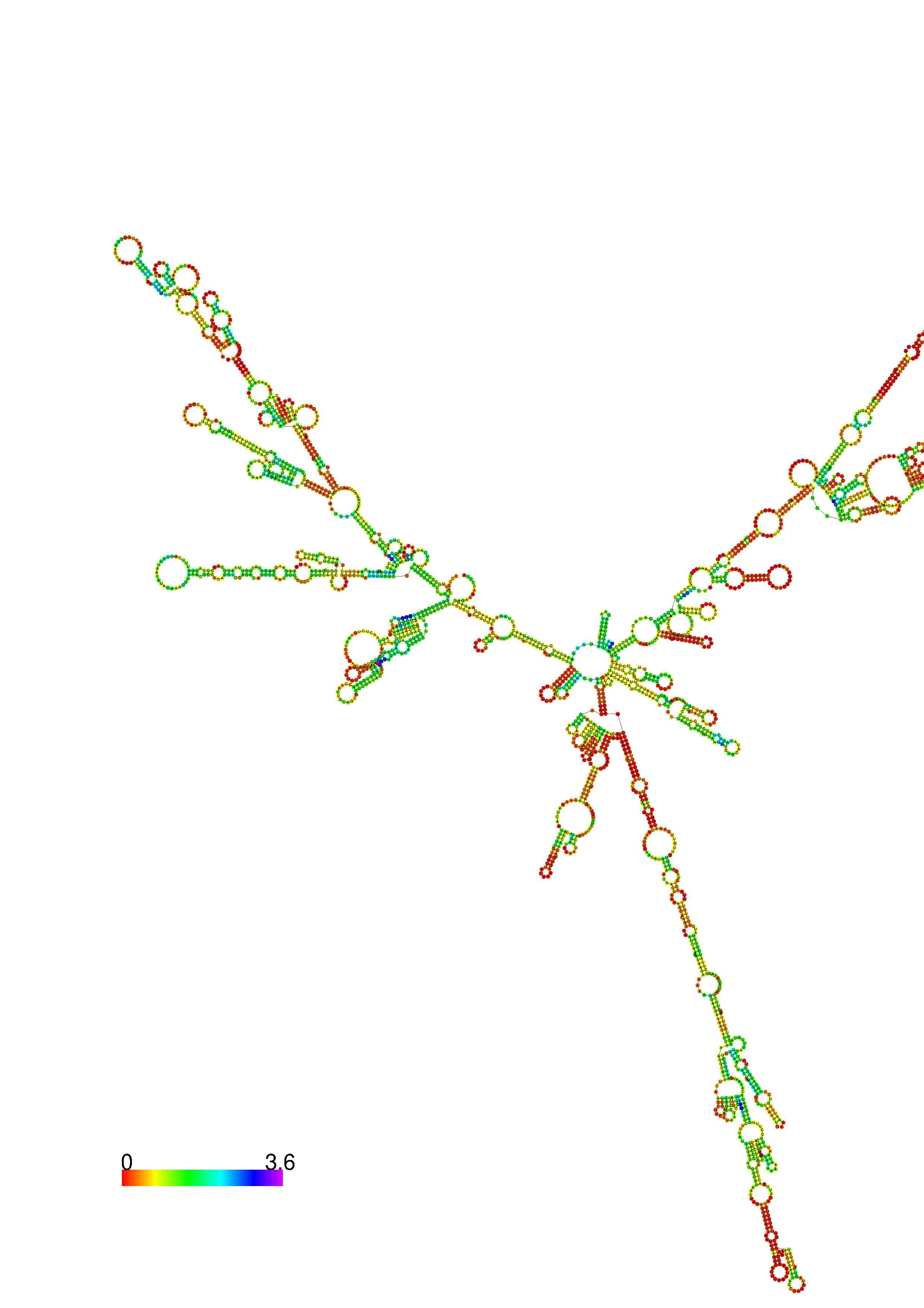
III. Graphical output
1. A note on base-pairing probabilities:--
The structure below is colored by base-pairing probabilities. For unpaired regions the color denotes the probability of being unpaired. The reliability information of RNA secondary structure
plot in the form of color annotation, Positional entropy ranging from red (low entropy, well-defined) via green to blue and
violet (high entropy, ill-defined).
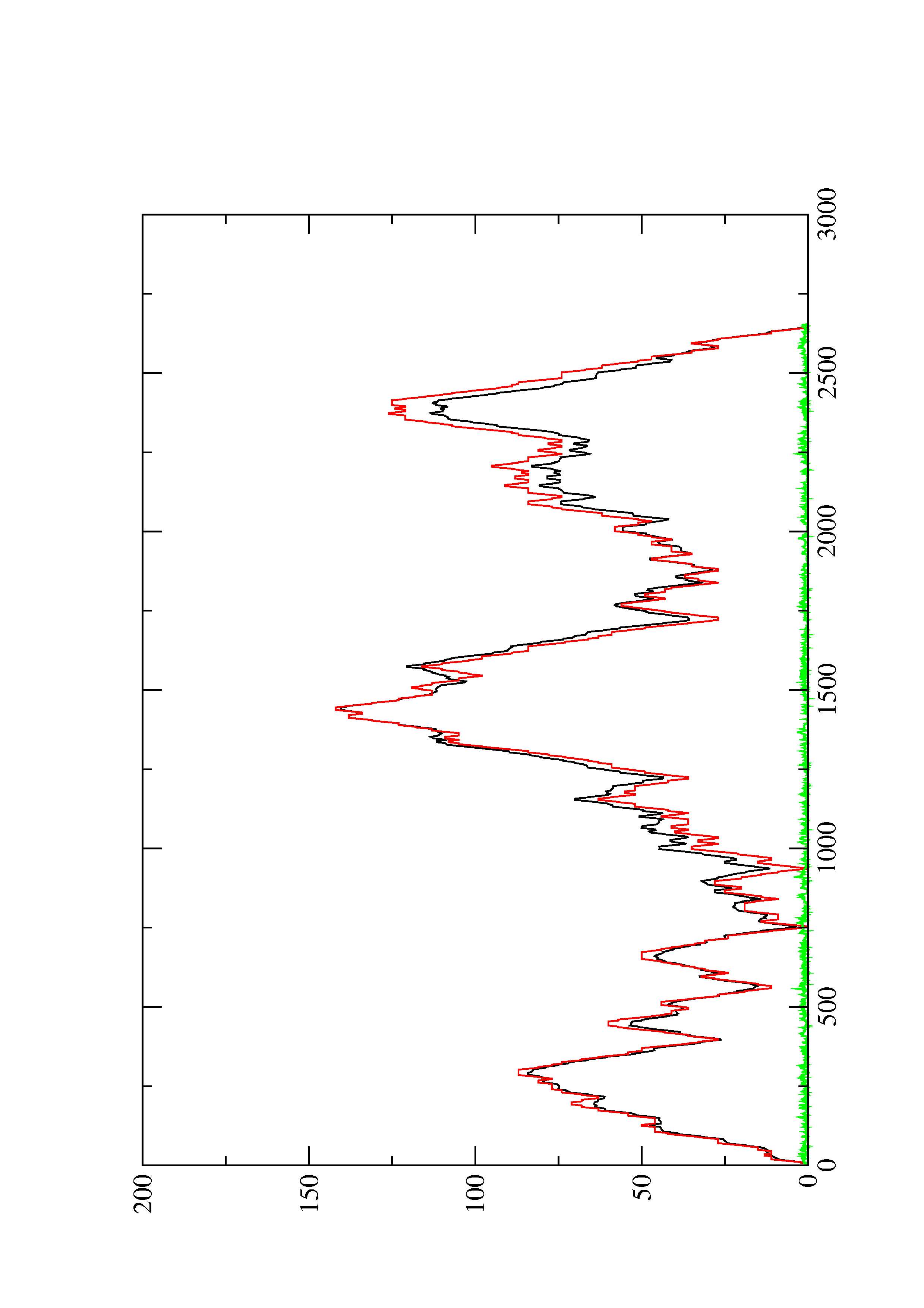
2. Here you find a mountain plot representation of the MFE structure, the thermodynamic ensemble of RNA structures, and the centroid structure. Additionally we present the positional entropy for each position. The resulting plot shows three curves, two mountain plots derived from the MFE structure (red) and the pairing probabilities (black) and a positional entropy curve (green). Well-defined regions are identified by low entropy.