lncRNA Structure Prediction
Species Name : Withania somnifera Sequence Id : GBHJ01023781.1
I. Result for minimum free energy prediction
The optimal secondary structure in dot-bracket notation with a minimum free energy of (-442.30) is given below.
1. Sequence-
AGGUACGUAACAUUACUACCAAGAGAUAGGUGAAAAAAAUAUUAAGAGUAAAUCUAAGAC
GUGCAAUAUCAGGGAUAACAUAUGACAGAGCCUAUCAAGCAUAUCAGAGGCUUAACCUUC
ACAGAUUAAAGUAUGCUAUUCCAACAGAAACCAUAUAUAAUCAACCAAGGGUUUACUUAA
AGCUCUGGACACUAGUAUGCAAACAGAACAGGUUCAUCGCAUGCUUCUUUGAAAAGGGAA
AGUAAAUGACGUAAUGGUUGCGCUAAAGCAAUUAGUAGGAUAGGGAGUAUACUAAGUAAA
UUAAACCAUAACUUAAUGAAGAGAAGGCAUACAUUCCAUCCAACGAUAUAUAGUAGUUUA
GAGGUCACAUUACUAUUGAAGUGCUCUAACUUAUCUACGACAGAAGGCUUAAGCCCAAGU
CAGCUUACACAUACAAAAGUACACAUGCAUACUAUAUACUUAAGACAUGUUGUAGCAUAU
AAUUAACAAUAGUCUCAUGCUAGUUUCAACAGGAAAAAAGGAAGAUGAAUGAGAUCGGAC
CGUUCACUUAACCUGACUCAUCCAGUGCUAAAACCAAAAAGAACAAAUAUGCAAGAGACU
UAAUCUAAGCAAAAUUAGAUCACCCCAGGCACUUGUUACCAUAAUUGGGAUAGCAGGGAU
GCAGUAAAAGGACAGAAGGUAGCAAGUUAUCCAUAUAUUGAUCAGAUUCAGUAUCAUGCU
AAAAAAAAUGAAAACUUGCACCUUGUUCAAACUUCAACUCAAUACAAAGUUAGCAGUUAC
UGCAGGCUUAAGCAAGUACAUUAGACUUAGAGAGUUAUUUCAAUGGUUACUGAAACCUAC
UUCACUUUUUCCCAGAUCACCUAAUCAAGAACAUAAAAUCAUAGGUAAUAAUAUUGAAUA
AAACAAAUAACCAGCUUAAUACAUUAGACAUUAAAUAACAAAAUUCAUGCAUGAAUACUC
GUAACAUAGACAGUUAAUUACAACAGACAAUUACUAAGUAUUACGCCAAAUCAGUGGCUU
GUUUUCCCACCCUAGAUGAGGAAAACAGUUAAAAAACACAGAACACCAUAACACACAUUG
CAACACCAAUAUGAAAUCAACGAAUUAAGGAAAUGAUUAUCAAAAUAAUUUGUUAUCUCA
CAGAAAAAUGGGAAAACAAGUAUAGGACAAGGAUUACACUUAUGCCCUAGAUUUCAUGAA
CAGAAAAAUCAACAUUUAAAUUGACAAAGCUUCUUAAUAUAGUUUUGCAAGGACAUUUGA
AAGUCCUAUAUCCCCAGCCCUUUAGAUAUUAAACCUUUGGAUUCGUAGGAGGUAGAUUGA
ACCCUAUGUCAAUCCACACUCCAAUAUAAGAUGUGGGUUUCUUUGCUAAAAUAGUGGAAG
CAUAUCAUCAAGCUCUAUCUAACUCACAUCUUACAAGUCCUCUUGUUGUCUGGCACUCAC
AUAGAUCCUUGUGCAGUUAAAUGAAGGAGGAAUGAGGUAAACAAGGCUCAAGAAUCUGUU
UACUUCUGUUCAAAACAGUCUUUAAACAAAAAAAAAACUCUUUCUAGAUCAUUUAGCAAG
UAGGUAGACCACACAAAGGAUGGCCUUUAAAACAAGAACAUCGGGUGGGCUAAGGGGGUU
UCAAUUUGCGAUUUGUAAGUGUCGAGAAGAUCAUUUCAAGAUCAGCUUGAAAUGGGGGAU
GAAUGGCAUUGGAAUUGCCUUUCUUAUCCUUCAAAUCAAAACGCAACCUUAAUGUCUCAU
UUUUCAUUUUAAAAAGACAAUAAUUUUCAAAAGACUUUGGAAAAUGAAACAUGAAAUGAA
AACAUUUUUCACACACAUGUAAAAAUCCAUAAUCACAUAACUGGUACACAAAUAGCAGAU
UAGUCUUUACUGGAUUGUUCUUAAUUUCUUAACAAACUCAUAUUUAAAGUAAAAAACACC
AUGAGUUGUACAAUGAACAAGAUAACAAACUUAAGCUAAGAACAACAUGAUGAAACAGUG
UACACAUCCACGAUAAACAAAUAGGUAAUUUCAAGAUAAUUUCAAGUAAAAAAAUGUGGA
GGUUGAGAGUGUACCUGUUCAGGAGAAGGAAUCAAAGCCAGGACUAGAAAAAUGACCACU
UGUUUCUCAUAUUGACACUUGUAGAA
2. MFE structure-
.((((.(((....))))))).(((((((((((............(((.....)))..((.((((......(((.................))).....)))).))...(((((..(((((..........((((((((.((((.........................((((((.....))))))))))...))))))))
.....((((((((((((.(((((....((((....(((((((............(((((((......)))))))((.((((.((((((((.((......(((((........)))))........)).)))).)))))))).))..........(((..((((((..((..((((((((((((((((......((((...
......(((((((((...((..((((..........))))..))((((.(((.((((.........)))).)))))))............((((((.(((((((....((.(((....((((((.((((((.........)))))))))..)))......))).))))))....................))).))))))
..((((((......))))))...(((..((...((((.(((....))))))))).))).((((((((..(((((..((((.(((((((.((((((((((((......))))))).)))...........)).)))))))))))))))).......((((........)))).....))))))))))))))))).......
.)))).....)))).))))))))))))...))..)))).))..)))))))))).....(((((..................)))))....................................................))))...)))))))))((((((.((((....(((.....((((....((.(((((((....(
((((((((......)))).((((((((((.......)).))))))))(((((.(((...(((((........(((.(((((((......(((((((((....(((((......))))).........(((((((.(((((........))))).))))))).((((.(((((......))).)).)))))))))))))..
.....................((.(((((((.........)))))))))(((((........)))))..(((((((((((....(((...........(((((.(((((((((.((((((........)))))).)).))))......(((((((((((.....(((......(((....))).......))).......
)))))))))))))).)))))(((((((....(((..........(((((((((..((((((((((((((((..((((((((((.((........))))))))))))((((.(((......))).))))..........)))))))...))))))))).....((....)).))).)))))).))).....)))))))..)
))....)))))..))))))......))))))).)))..((((.((((((.(((((((.......((((((..(((((((((((.((((.......)))).)))..))))))))..))))...)).......((((.((((((((((((((.((((........)))).)))).....)))))))))).))))))))))).
....)))))))))).........(((((((((..((.((((((.((......)).))).)))))..))).)))))))))))...))).))))).(((((((...................)))))))(((((.((..((..((......((((...))))......))..))...)).))))).................
....)))))....)).)))))))..)))).....)))....)))).))))))))))))))....))))).....))))).................)))))))))))...................
You can download the minimum free energy (MFE) structure in
II. Results for thermodynamic ensemble prediction
1. The free energy of the thermodynamic ensemble is [-480.36] kcal/mol.
2. The frequency of mfe structure in ensemble 1.5046e-27.
3. The ensemble diversity 447.84.
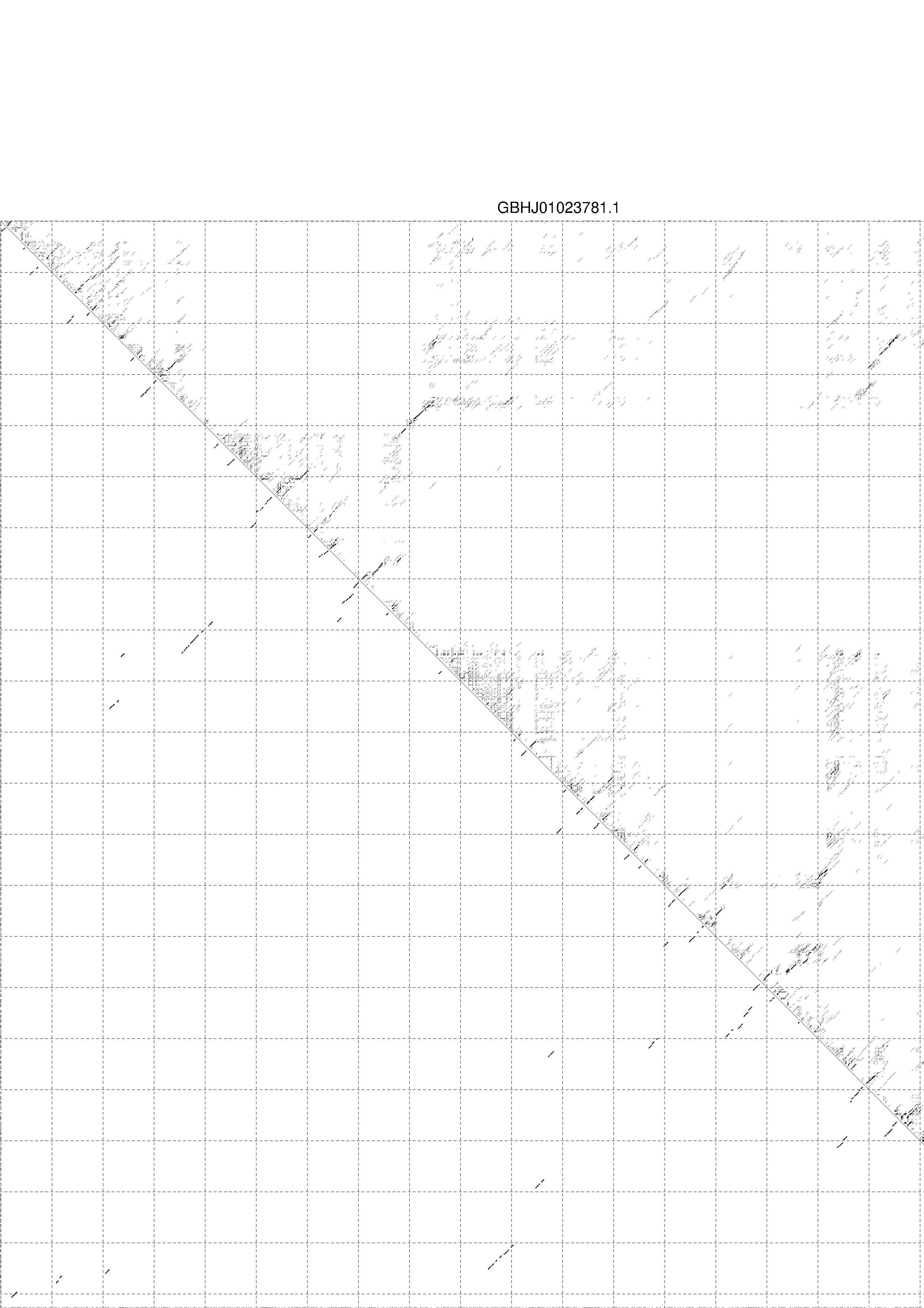
4. You may look at the dot plot containing the base pair probabilities [below]
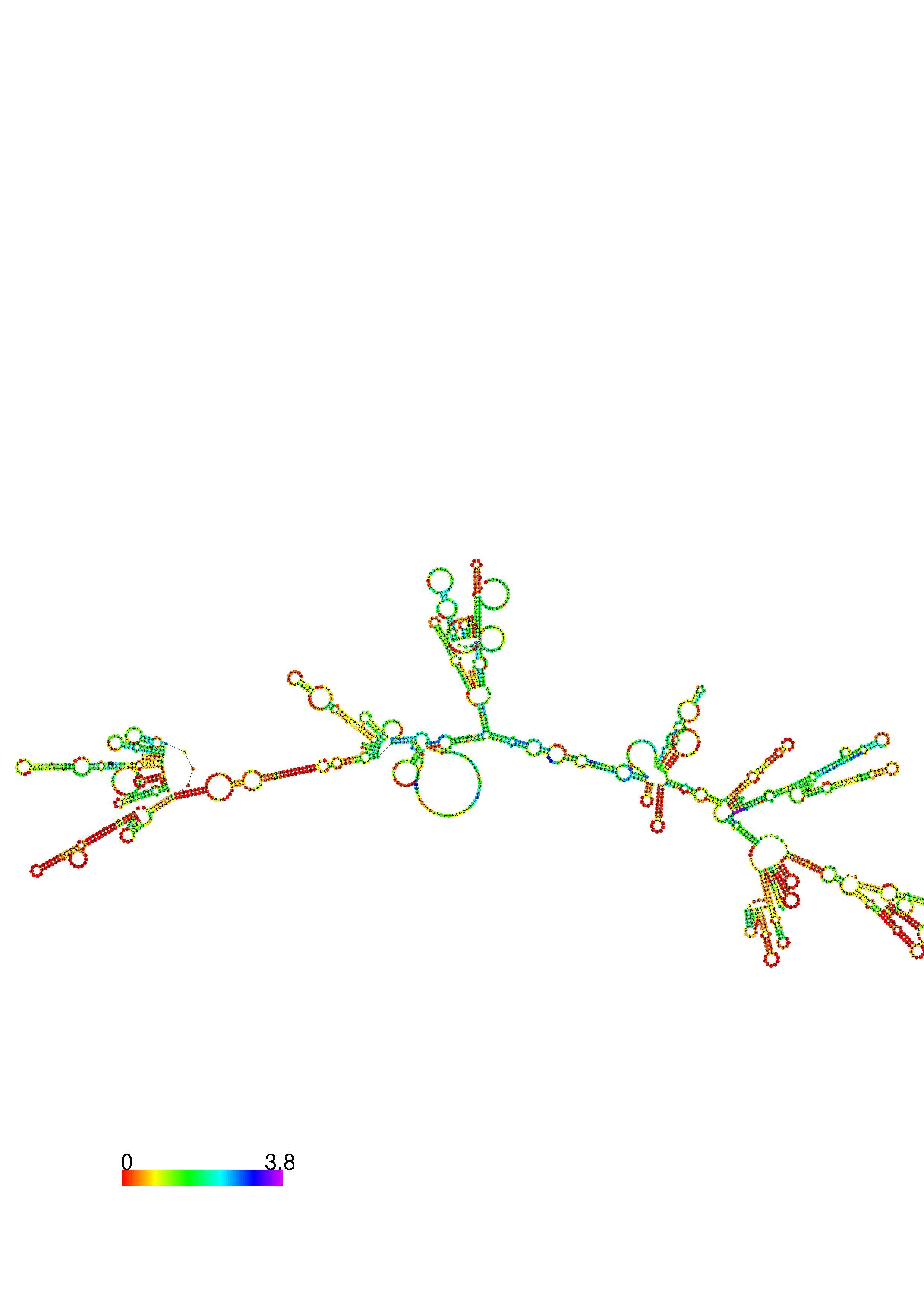
III. Graphical output
1. A note on base-pairing probabilities:--
The structure below is colored by base-pairing probabilities. For unpaired regions the color denotes the probability of being unpaired. The reliability information of RNA secondary structure
plot in the form of color annotation, Positional entropy ranging from red (low entropy, well-defined) via green to blue and
violet (high entropy, ill-defined).
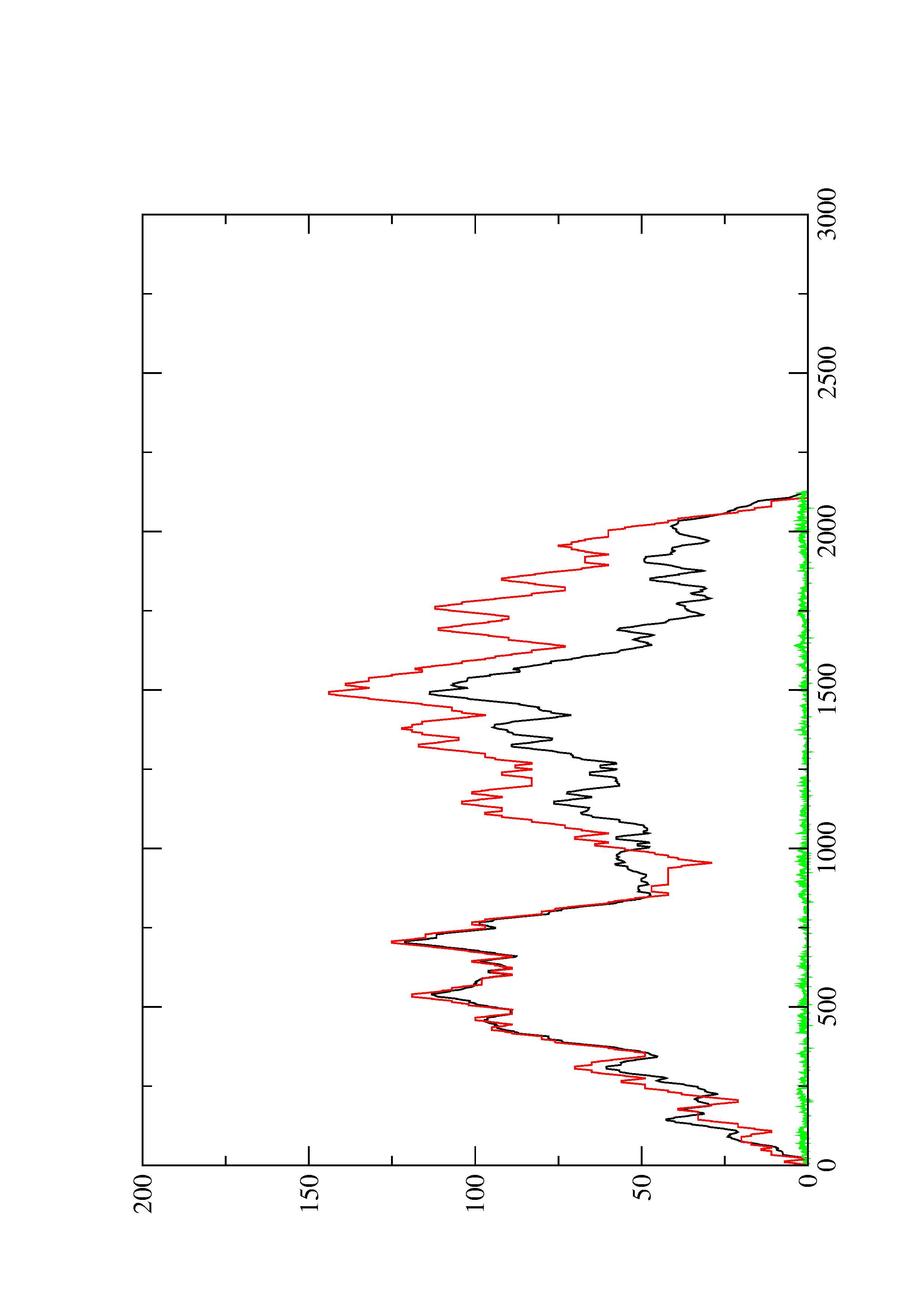
2. Here you find a mountain plot representation of the MFE structure, the thermodynamic ensemble of RNA structures, and the centroid structure. Additionally we present the positional entropy for each position. The resulting plot shows three curves, two mountain plots derived from the MFE structure (red) and the pairing probabilities (black) and a positional entropy curve (green). Well-defined regions are identified by low entropy.