lncRNA Structure Prediction
Species Name : Withania somnifera Sequence Id : GBHJ01024315.1
I. Result for minimum free energy prediction
The optimal secondary structure in dot-bracket notation with a minimum free energy of (-465.80) is given below.
1. Sequence-
UUACUAUGUGGGAUUCUGGCUAAACAAUUAUAGCAUCAGUUCAUAAUGUUAUGCGGUUUC
CUUGGUUUUUGCUAGCUAGGACAGACAACCACUUAGCUCAUAUACAAUCAGGACUAGACA
UAAGAAACAUGAUUGUCUCAAAGUUGGUUGUUGAUUGAUAAAUAUACUUUUGUUUAUAAA
AACACUUAAGAUGAAAUAUGCAUUUGAAUCCUGGAUGUUGGAACAACAUUGUUAAUGUGA
CUAGACAGUCCUUUCUCUCUAUCUUCUUCUUCGUCUUUGUUUGGUUUGUUGGCGUUCUUG
UCUUUAUUCUGAUAGGCGCCAAUUUCAACCACAUUCCAGCUUCUGAAAGAAGUGAAAAGU
AAAACGGAUUUUUUUCCUCCUCUUCAUUGUUUUCUCUGUCCAGAUGUUAUGAGGAAUUGG
UUCUCAUGUUUCUGUUAUCAUGAAGAGAAGAUAUAUAAUCACAUGUAGAAAAGAAAUAAA
AAGGAAAAUUGAGUAGUAAUCUCAGCAAAAGUGAAUUUGCUAAUGAUUCCAGAAUGGUAU
CUUUGCAGUUUCUCCAUUAUGUUUUUAUUUUUUUCAAGUUUAUCACAUCUAAUGUGUUUA
UAAAGCCAAUCCCACUAAUUUGGGUUGAGAUUUAUAGCAUGUUUAACCAAGCUUUCAAAU
UCUGCUUAUUCUGAAAAGUGUUUUUUGCCGGUAGUUCUUUUAGAAAGAGUACCUUUGGAG
AAUUGACUGAUCAAUUUGGAAAACAACACUUUUAGUAAUAUUAAAGUGGUACUUUGUACU
UUGUGCUUGGAAAACAACUCUCUAAUUUUGAUGCCGCACCAUGUCGGAUUCUACGAAAAU
ACACUAUUGGAGAAUCCAACACGCACACCUGCUUGACAUUUUUUAAGAGUCCAAGAAGCA
UAGUUCCGAAAUGCCUCUAGAAAAAACCAUUUCUUUUAAGCUUUUGAAAUGUUGCUUCAG
CUACUCAAGAGCACUUAAUUUUCCCCCCUAAAAGCUUGGCCAAACUAGCUAUUAGUUGAU
UGAUUCAUAUGUAUGUUUUGGUGAUUUUCAUCUUCCCGGAGUGGUUUUCAACCUUGCCCG
UAAUACCUAGAACGACCAAUUAAUGUUUUAUAUGAAAUGAUUCUGUAAUGACUCUGACCU
GUAUAUGUGGAACUACAGGCAGCAAAGGAGAAAUACAACUGGAGAUGUAGAGCAAGAGAU
AAAAGGUGGUCAAACGCUUAGUUUCAUAAUGUAAUUCGUAGCAGUUGGUAUACCAUGUAG
UCUUAUUUCUGAACCGCCUAGUUAACGAGACUUUGUUACGGGAAUUGCCUCCCUCAGCGC
CUAUCCACCCGUUUUCAGAUUUUGUAAACUUUGCUCUUGCUGUCUGAUUUUACUUUUUGC
CCUAUUCUUCAAGCCUUGAUGAUGGAAUACAGUUUUUCUAGCUUUGCUUAAUUAUGACGG
AAAUAUGGAGAGAGUGAAACAAUUCCUUAUUUUUAUAGAUGGAAAUGUCUGAUUGGAGGA
UGGAGUUAUUGAACUGAGUUUGAAAAGGAUGUUUAGGCAGGGGUUCAAAUUGAAUUGUGU
UUUGGAGUUCAUGAAGUUCUUGCCAAACCGUGUCGUGGAAUAUGUUUUAUCCGCAUGUUG
GUAUGUGCGGAUAAUAUGGUAUGUGAGGAUGUGAUGCUCUCCAUCCUCCAGGUACAUAGC
AAACUUUUUAAUCGUGUUCCAUUUUGGGUGUGGGGGUCAUAAAGAACUCCUGAAAGAUAU
UGCAGCUUCAAUUGUACAGGAUCUUCAAUUUUGUUAAUUUUGUUUUAUCGUUAUAUAAAG
UAAAUCAAGCUGACGAAUUGGACUCUAUUUGUCCACUGUAUUUCGUGGUUUGCCAAAAAU
UAGUACACUUGCCAGAUGCAGUUACUUUGAGUGCCG
2. MFE structure-
.........((.((((((((.((((.(((((((.((((.((((((((((((...((((..((.(((..((((..((.(((...((.((((((((....((...((((((((....(((((.............))))).....))))))))....))(((((((......)))))))...............((((((((
(((.((((((...(((((((...)))))))........((((....)))).........((((((((.((((((.......(((((.(((((((..((((((........)))))))))))))...))))).............(((((.((((((.((......(((......)))..)).)))))).)))))......
((((...((((((((.....))))))))..)))).....))))))))))))))(((((.(((((.((((((((((((((((((((((((((..((((((..(((.......)))..))))))..(((.(((...))).)))....)))))).)))......))))))))))))).............))))))))).)))
))....(((.((((......))))))).((((.(((((..((((..(((((((((...........((((((((((.(((((.(((.((((((.((((((...))))))))))......(((((((....))))))))).))).))))))))))).))))....((.((((.((((.(((.(((.((((((......(((
(.(((....((((.((.....(((.(((((((.(((..........))).))))))).))).(((....))).)).))))..)))))))))))))..))).)))..)))).)))).))(((((......))))).....((((((((...((((...))))...))))))))..................))))))))).
.))))..))))).))))....)))))).)))))))))))(((((...)))))......)))))))).))..))).))..)))).))).))...))))...))))))....)))))).)))).)))))))....(((.((((((..........))))))))).......((((((((..((((...((((((...(((((
...(((((......))))).)))))......(((((((((((..(((((.(((.((((((.........((((((.(((...((((((....)))))).((((......)))).....(((((..((.((.(((((((((((.((.......(((((..((.((((((((((((((((.((.((((((((((((...((.
..(((((........)))))...)))))......))).)).)))).))))))))))).(((.((((..............)))).)))..)))))))))..)))))......)).))))))))))))).))..))))))))))))))(((((((((((((((((((((((((((.....((.(((.(((.....((((((
((((..((((((((((((....))))))))))))..((.((((((((((((.((.....))))))))).....))))).))...........))))))))))....))).))).))))))...)))))))...)))))...)))).)))))))..(((((((...((....))...))))))).......))))))...)
)).)))))))).))))))))..))))))...)))).))))))))...))))))))......((((.(((.((.....)))))))))..)))).)).
You can download the minimum free energy (MFE) structure in
II. Results for thermodynamic ensemble prediction
1. The free energy of the thermodynamic ensemble is [-500.22] kcal/mol.
2. The frequency of mfe structure in ensemble 5.58792e-25.
3. The ensemble diversity 345.61.
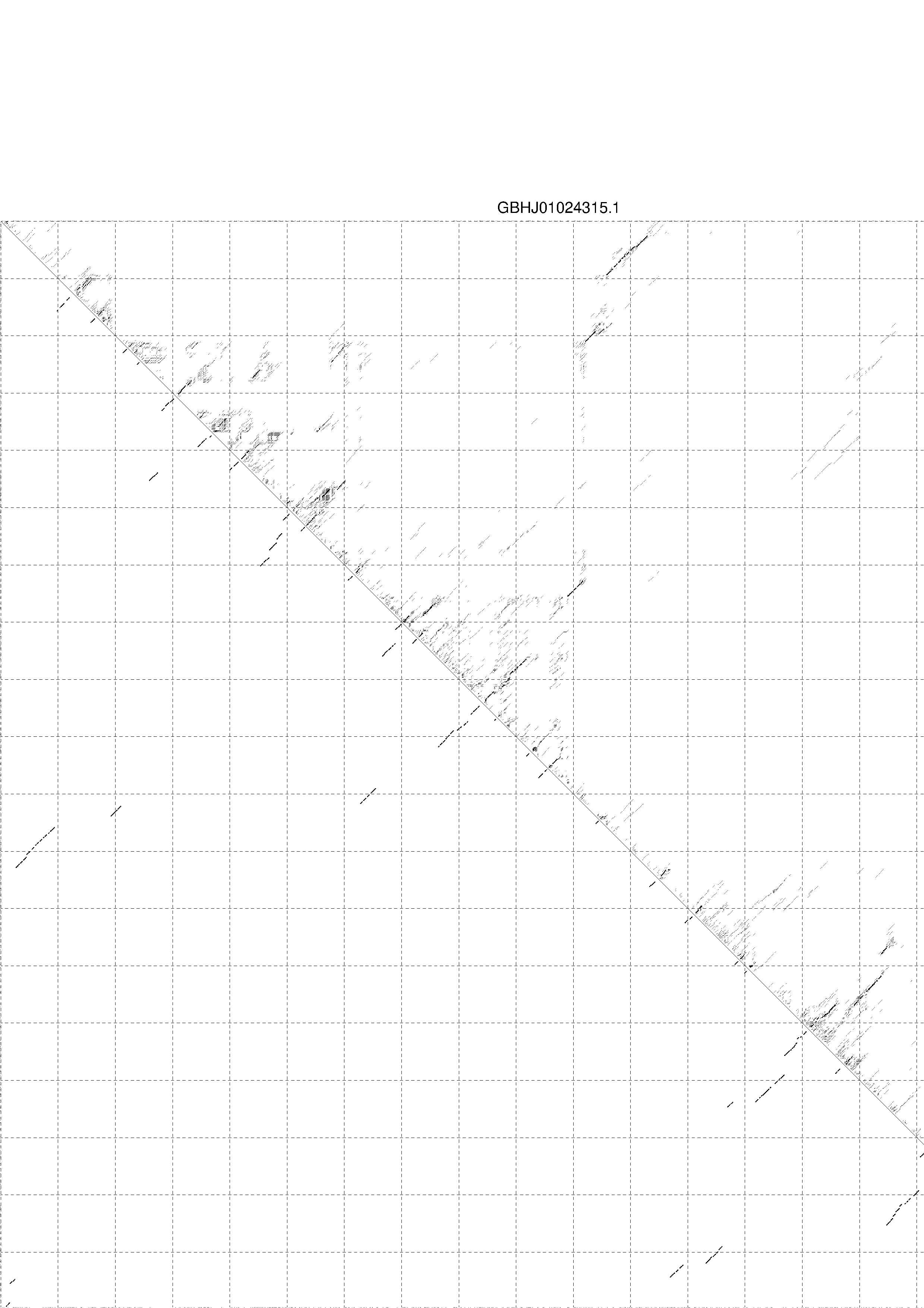
4. You may look at the dot plot containing the base pair probabilities [below]
III. Graphical output
1. A note on base-pairing probabilities:--
The structure below is colored by base-pairing probabilities. For unpaired regions the color denotes the probability of being unpaired. The reliability information of RNA secondary structure
plot in the form of color annotation, Positional entropy ranging from red (low entropy, well-defined) via green to blue and
violet (high entropy, ill-defined).
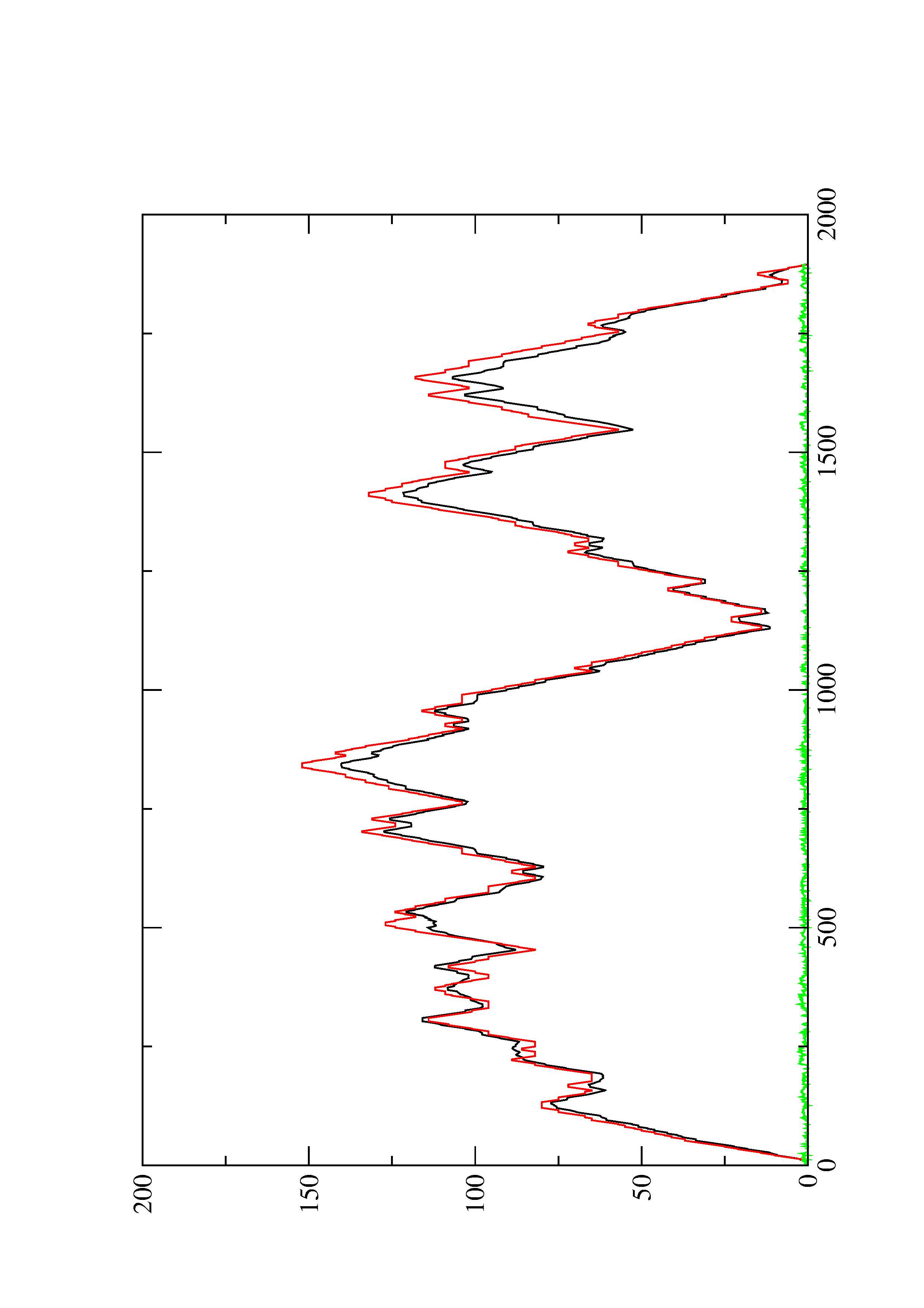
2. Here you find a mountain plot representation of the MFE structure, the thermodynamic ensemble of RNA structures, and the centroid structure. Additionally we present the positional entropy for each position. The resulting plot shows three curves, two mountain plots derived from the MFE structure (red) and the pairing probabilities (black) and a positional entropy curve (green). Well-defined regions are identified by low entropy.