lncRNA Structure Prediction
Species Name : Withania somnifera Sequence Id : GBHJ01024426.1
I. Result for minimum free energy prediction
The optimal secondary structure in dot-bracket notation with a minimum free energy of (-684.60) is given below.
1. Sequence-
CAAGAGUAAAAGUAGGCUCAUUUAAAUAGCUCAACUUAAAUAGUUUUUCACAAAAGAUAA
GACAGUAAAAAAUAUUCUUUUAUUUUUUUCUCUUAAAACAUUCAAUUUUACUUCUGUUCG
AGAGUUCCUAUAAUUGAAAUACCUUAGCCUUUGCAUGAAAUCUACAUAUUGCCCCAUGUA
GGGGGCCUCUAUUCCUUGUCAAAGGGUAUGAACCUAGUUUCUCCUAUUGGAUCCAACUAG
UCUAUCAAAGCCAUACUUAUUGGUCCAAAACACAUAGUCUUAGAUUAGGCAACACAGCUG
UAUGGGUAUACCAUACUUAUGCGCACCAAUGGUACUAAUACACCUCCCAUAAAAUAUAGU
UACUUUGCUUAACUUUCAUUAUCUCAUGAUUAGUGGAAUUCACCUCAUGGGGAAAAACUA
UCAAGAUACAAUUUAAUUAAAUAACACAAAUUGCGUAACUCAAUCAAUUAUGACUCUUCA
UUAACAUUUUGCUUCUCAGCUUAAUACUUAAGUAUAAACAUCAUUACUUCUCUUCAUUAG
GCAUAAUUAGCAAAAAAUAUUUAAAUUUAAAAAUGCUUCAUAAAUAGGUUUAAAUCCAAG
UGAAAGUCAUCAAUUAUGACACUUCAAUUCUCAAUUCAAGUUUUAAUAAAAAAACUUUAA
AGGAAAUUCAUACUUCAAGACUUCUAUCAUACCUUCGUUUUAACUUUCAAAAUCAUGCUA
ACCCAUGAGGACUUACACAAUUUAUCAAAUAUCAUGUAAAAUUCAUAAUUAUAACUCAAA
UUUAUCAAUACACCAUCAAGUAAGGGAGUCUUAAUAUUUUAAUAGAAGUAAGAACCCUAA
CAUGCUUCUACAUCUUCAAACCCUUUAAGAAAAGGGUUUUUUGGGCACAAGGAUUGAAGG
GAUCCUUGUUCAUAACCCACGUACUUCAAUCAAAGAACUUUUAUGGAAGACAUAAAAGAA
AAAAAGAGCCCCUCUAGAUGUUUCUAUUGAAACCCUAGUUUCUUACUUGAAGAGGAAUUU
GAAGAAAAACCUUUUUCUAAUCAAUAUUAUGUAAAGAAUGAAAAGUUUAGGGUGUUUGGG
GUUUAUAUAUUGUCAUAUUACAAGUCCAAAAUGACAUGGGUUUAAGUUAAAAGAGCUUGA
UAACUGACCAAAUUACUCCUCAUGAAACAUACAAGAACACAUCGGGAUCACUUCAUGAGU
CACUUCACGACUCGUAAAGUGCCACUCCACGAAUCGAGGAGUGAUUCGUGAAACAGGCUU
UGGCUACUAGAAUUGGAAGCCUGUUCCACAAGUCAUACCAAAACUCAUGGUAUGCAACUC
CAUGACUUGUGGAGUAGUCUCGUUGAAACUAAGUUGAUGAAUUAAAUUUCUGGAUACUUU
GCACAAUUUAUGCAGUAGGGCGUGGUCCUCACUUCACGACUCAUACAAGGGACUUGUGGA
AACCUGACAGAGCAUGAAAAACUCAGGAACAUUAGUAUGGGUACAUUCACUACUCGUGAG
GUGAAACCUUGGAAAAUUAGGACUGAUCGAGCCCCCUCCCAGAUCAAAGGAAAAGGGGAA
GAAAGGACUUCUUGAGAAUUAGGCUUAGUUGAGCCUCUCCAGCUCCAAUAAAUAGGUCAU
AUCCCAUCAGAUUAUGCAAAGGAGAUUCUCGAACUAGGAAAUUAGACAUGCAAAGAUUAG
AAAUGGAUUAGGAAAAAUAAGUGACUAAGUAAAGGAAAUAAAAUUUACCACCUAUUGCAA
AUAGUCACAAAGGAAUAACUGAGGCAUAAGACAAAGAAUCAGUUGUGGGCCGAGCCCAAU
ACAAAUGAAGAAUAUCAAUUGUUCUUAAGGGAAGACAAGCAAUAAGGCCCAACAAACUUG
GGGAAUGUUGGGCUUGAACCAUAUAAAUACAUUCCCGAAUGGCCUAAAUCAUCCUACACC
CUAAGGGGCCUGACCCAAAAGAAAGUACUAAAAUGGCUUGAAUUCCAACAUAGAAACACA
UAGGCAUCAGUUAAGACAUAGGCAUCAGUUAAGACAAAAAUGCAAAUGAUCAGUCUCUUA
AAAUAGAAAAGCACGGGUUCCUUUAAAGGGUUUGAAGAGAUGAGCUACUGGACAUGCUAU
AAUAAACCAUUUUUAGGGAGAUGAUUAAGCUAUUUAAUCUAAAUAUAUGCAGUCUCAUAA
AUGAAUAAAGGACAAACAUGCUUGCUUUAACUGAAACUAAUUUGAAAGAAUAAGGAAAAG
GUAUGCAUAAUAAACUUGAUUUUAUAUAGAGCUAACAAGAUCAAGAGUUGGUUAUGACUC
UCAGCCAAUCAACACAUAUAGGUAUAAUGGUAUUAGCAAAGGGACAACAAAAGGCAAUUG
ACAACAUAUCAACUAUCAUGAUGCUAGUCAUGGAUAUCAUAAGAUACUGAAUACUAAGAU
UAGAAAGGUACCUAGACAGUGAUUAUGCAUCACCUACAUACACAUCAAAUGGAACCAAAU
AACAUAUUCAUCCAAAAUAAUUUGAGCAUCAAGAGUAGGUUCUUAUAUCAAUCAAGUAGG
CAGCUAGGUAUCAGAAGCUAGGUAGAAACUAUUAGGAAUAGAACUUAAGACAGGUGAUGC
AUAUUAAUAUCAAGUAAUUUUAAGCAUCUUCAAGGCCUAGUAGAGAAGCGUGUUCUAACA
GAGAUCCUUUAAAGCCAAAUAAGGGAUCAUUUCAACCACAACUUGAGCACAAAUUUUACU
CAACAUAUACUUUUCCAAAUGACUAAUUAUCUGUCAUAAUAUACAUCACAAGAGUUUGCU
AGAAGACGAUUCAUAUUAUUUAAUGCAAAAAAGACACAUAUUAUAGGCAAUAGCAUGCUA
GUUUAGGAAUAGUUUGGUUGAGGCUCAGGCAUGUAGAGAUAAGAAGAGAAGGUAGCUCAU
UACUAUAUGUUUUAUUUACUAGCCAUAAACACAAACAUGCUCAUAGGGAUGUCAAUGCUU
AGGUUAUUUGAAUGUGACUCGAAAAACAAAAAAAGAAAGCAACAUAUCAUAUAUUUCAUA
AGAGAAAAGAAGAAAAGAGAUGCAUUUAACUUAUGGUCAUGAAUUCAUGAACCCCUUAUC
UAGCAUAUGCAGACUACUAUCUUUAUCAAAUUAACAUAAUUAUAGGUAGAGGAUAUGUAA
GAAAUAUCAUCUACUUCUAUAGCUUCAAUUAUAG
2. MFE structure-
...((((........))))........((((.((((.....))))..........(((((((.((((........((((((.(((((((((((((..((((((((....((((..((......((((((((...(((..((((((...((((........(((((((...(((((......))))).((...(((((...
.)))))...))(((((((((((....(((((.((((..(((.............)))..)))))))))......(((((((((.((((........((.((((((((((...))))))))))))......(((........)))..............((..(((((((......((((((((((((((...(((.....
))).))))))).................((((((.............))))))((((((.((((.((((((((((.((((((....(((((((.....(((((((....(((((..........)))))......)))))))......)))))))...(((((((((((...(((...)))...)))))))))))...))
)))))))))).)))).(((....))).((((...((.(((((((......))))))).)).))))...................(((((((((((..(((((.((((((((.(((((......)))))....(((((..((.........))..))))).((((((.................................(
(.((((..(((...((((((....))))))))).)))).))..((((.....(((((((((((((((((....(((((...(((..(((((((((.....)))))))))))).)))))...................((((((((.....)))))))).......((((((..((((..(((((....))))).))))((
(((......((((((...((((((((((....))))))..))))...(((((...((((...((((((((((((((((((.(((((...(((((......((.(((......))).))((((((((((.....))))))......)))).......((((...................((.((((((.(((((((((((
((((((.(((.((((..(((...)))..)))).))).)))))))))))))))(((((((((..............)))))))))((((((((((((((((.......)))))..........))))))))))))).)))))).)).....((((....(((......))).....))))((((.......))))((((((
(.(((((((((((((((((.((.((((((...))))))))..(((((...............))))).....(((((((....)))..)))).)))))))))))..(((.........))).((((((..(((((((.((........))...)))))(((((.....)))))..........((((((((((..(((((
.((...........((......))...........))...)))))..))).)))))))...........))...))))))....................(((((((.(((((((...............))).))))...)))))))......((((((((................)))))))))))))).))))..)
)).....(((((((.....)))))))..)))).)))))...))))).))))).....((.(((((((((((.......))).......)))))))).))................)))))))))(((......)))...................))))..))))..)))))......(((..((((..(((..(((...
.......)))..)))....))))..))).....)))))).....))))).....)))))).)))))))))))))))))...))))....((((.(((((((..........((((((.(((((.......)))))..)))))))))).))))))).....)))))).((((.(((......)))))))............
))))))))..)))))..)))))))).))).....(((((((((.............))))))))).)))))))))).......(((..............))))))))))....)))))))..))............((((.......))))....((((((....))))))..((((....))))))))...)))))))
))....(((((((((...(((((.....)))))((((.............((((((..........((((............))))...........))))))............))))....)))))))))))))))))))).....((((((((...(((.(((((((....(((((........))))).....)))
))))..)))......))))))))((((.....))))......(((((((((........))))))))).............(((((((..........((((((....(((.((((..(((((.........)))))...............(((.((((((((.....)))...........(((....((........
))....))).))))).)))......)))).)))...))))))))))))).))))))).....))))..))))))..)))......(((((((..((((.......))))...)))))))...))))))))......))..))))...))))))))...................((((...............)))).))
))))))))))).))))))................(((((((....)))).))).....(((.((....))))).)))).)).)))))................(((((((.(((((.......))))).)))))))....))))..........
You can download the minimum free energy (MFE) structure in
II. Results for thermodynamic ensemble prediction
1. The free energy of the thermodynamic ensemble is [-746.47] kcal/mol.
2. The frequency of mfe structure in ensemble 2.54104e-44.
3. The ensemble diversity 724.22.
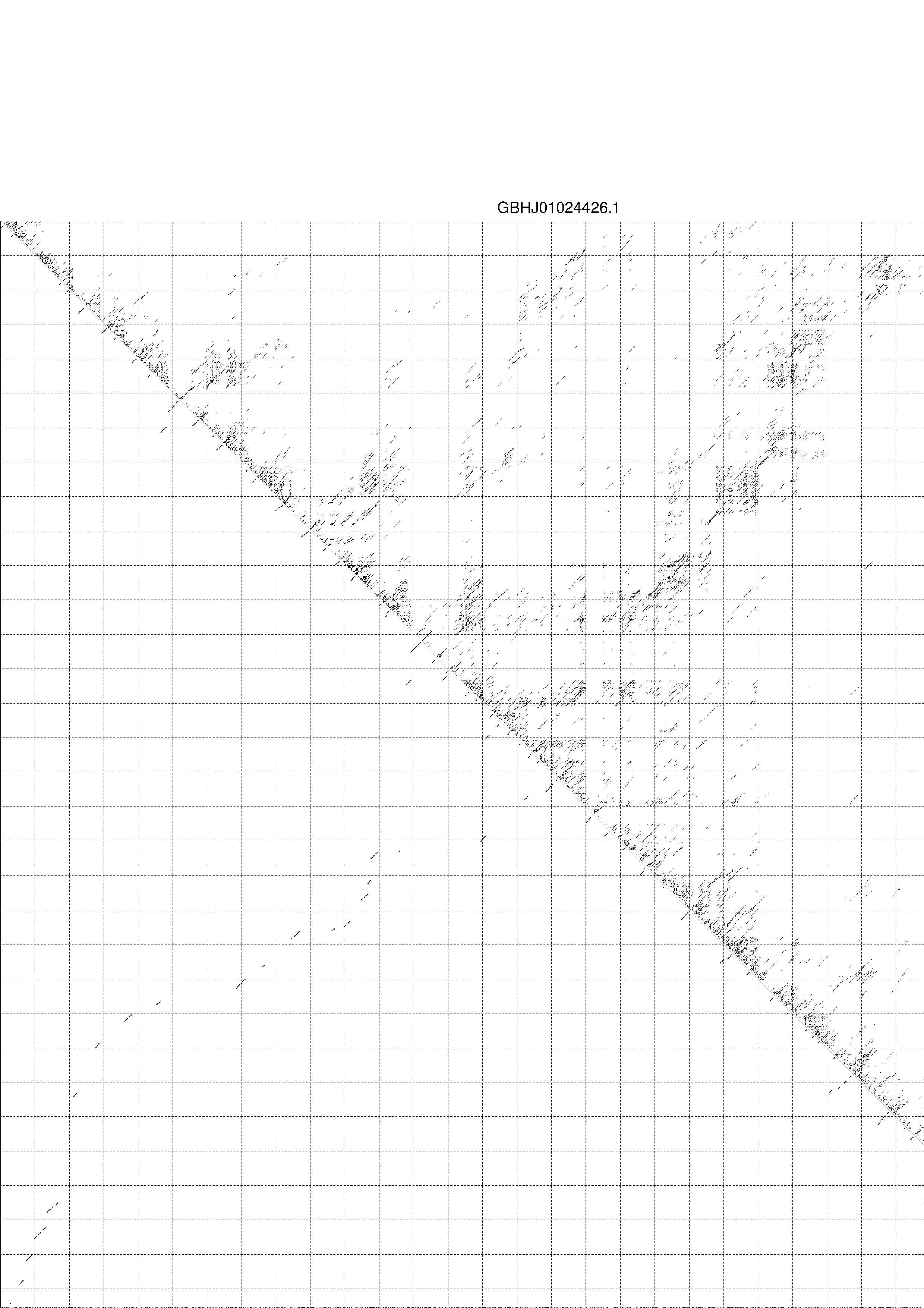
4. You may look at the dot plot containing the base pair probabilities [below]
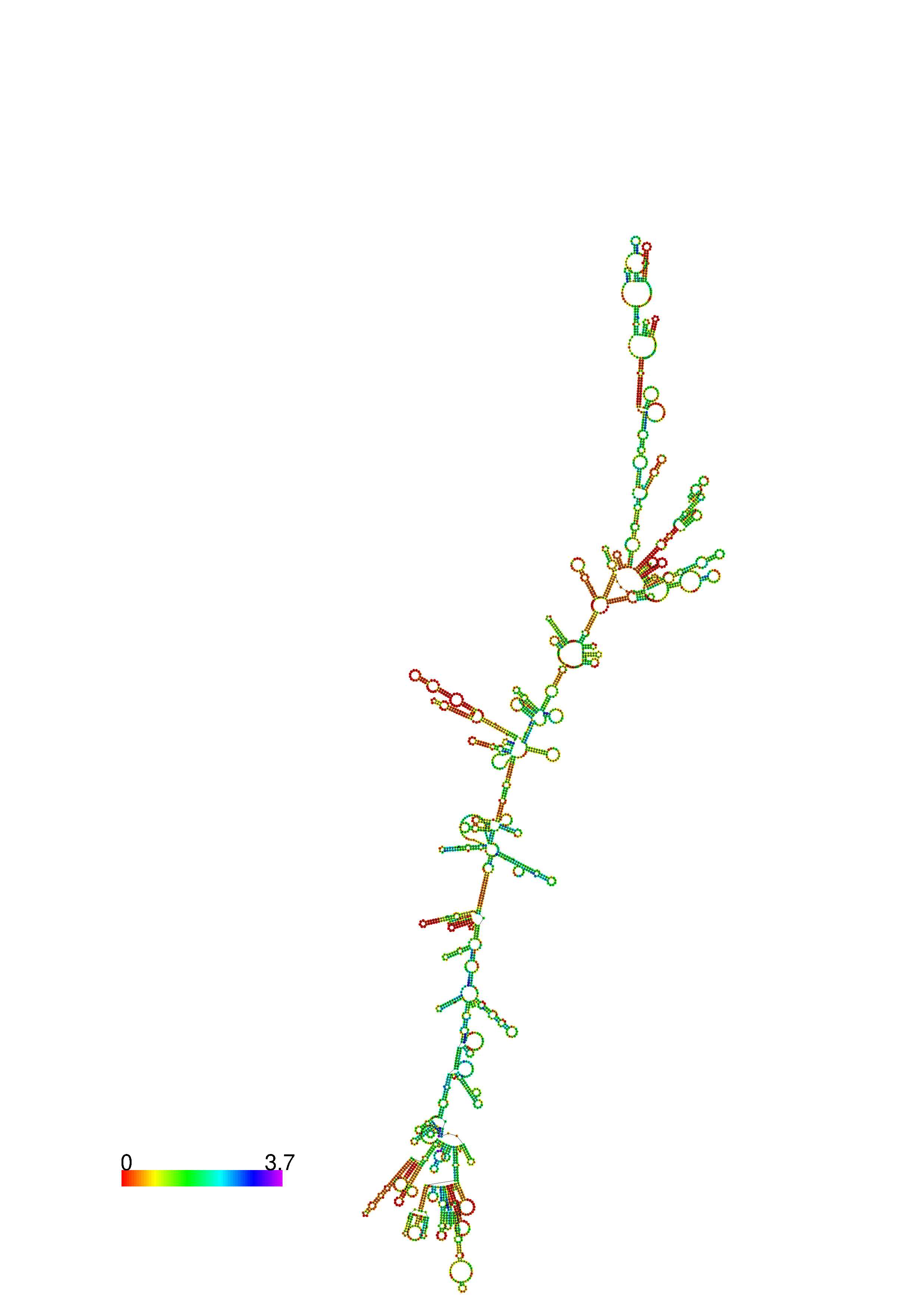
III. Graphical output
1. A note on base-pairing probabilities:--
The structure below is colored by base-pairing probabilities. For unpaired regions the color denotes the probability of being unpaired. The reliability information of RNA secondary structure
plot in the form of color annotation, Positional entropy ranging from red (low entropy, well-defined) via green to blue and
violet (high entropy, ill-defined).
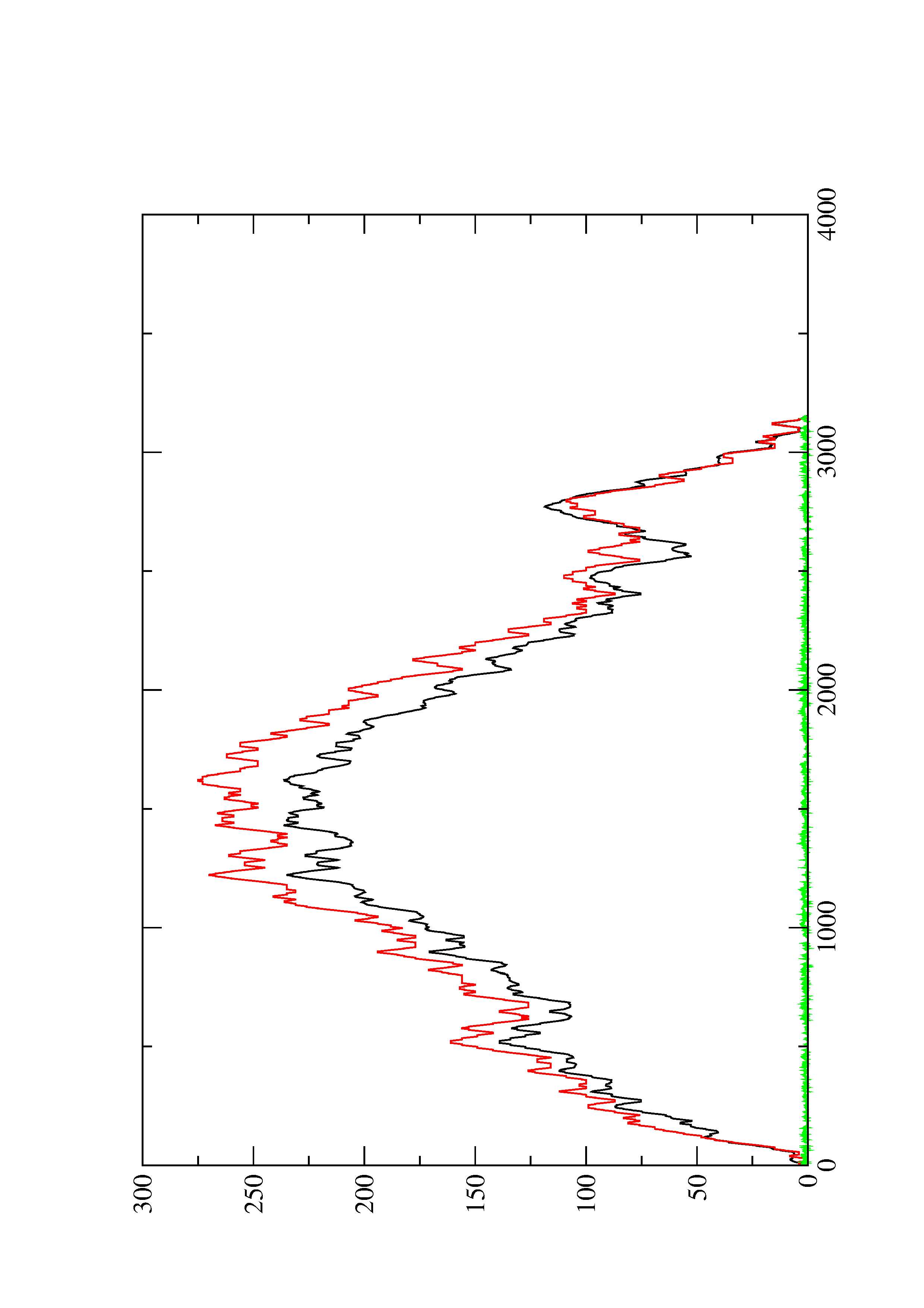
2. Here you find a mountain plot representation of the MFE structure, the thermodynamic ensemble of RNA structures, and the centroid structure. Additionally we present the positional entropy for each position. The resulting plot shows three curves, two mountain plots derived from the MFE structure (red) and the pairing probabilities (black) and a positional entropy curve (green). Well-defined regions are identified by low entropy.