lncRNA Structure Prediction
Species Name : Withania somnifera Sequence Id : GBHJ01024765.1
I. Result for minimum free energy prediction
The optimal secondary structure in dot-bracket notation with a minimum free energy of (-590.01) is given below.
1. Sequence-
AAAGAAUUCUAGAUAUACAUAAAAGUUGGGCCUUCAAUGAAUAUUUACCUUAAAUUAACA
AAGAAGCCCAAAUAGCAAGCCUAUUUAGAAAAAUUGGCGCCUCAUAUUUUAAGUUUCCCG
CGAAAGUAUCAGCCACAAUGCUCUGUGAUCUCUCCGGCGAAAAAUUGACCGUGCAGGAAU
UGGAGAGCUACCGUUUGAAGGAAAAAUCUCCGGUAAAUCUGAAGGUGUCAGCGCGACGCU
UGCCGGAAUUUCUGCAAUUAAAGGUGAAGCUGCCGGAGAUGAUGGAGUGAGUGCCGGUGA
AGAGGCUGUCAGUCGGUGGAAUUUAUGCCGGAGAAGGCUGCUACACUAUAAAAUCGAGGC
UGCCAAUUCUAUGCUUUUAGGCCACUACACACUGGACAGAAGGCUGCUACACUAUAACUA
UAUGAGCUCCAAAAUUCAGCUUCACGCCACAUGUUCUAUUGUCCUUAUGAGCUCAAAAUA
GAAACAGAUGAAAGCUGCUACUCAAUUUUGAUAUAGGCAGGAUCACACCUGCCAUCUUUU
CUAUUCAAGCAGAGAACACAAACUGACUCCAAGCCAUAGGUGCGGAUCCUGUUGGUCUGC
UCUCUCAAAAUGACCUCCAUCAUGCAUUAGCCCGACCAUCUAACUGGUACCCAAUAAAAU
CAGACUCCAUGCGUCUCUCUCAAGUCCAGAAGGGCCAAUACAAUUCAGAGUUACCAGUAA
AUAGAGGAGGAGCAAGAUUUAACUUUUCCAAUGGAAAUUCUCCGAGCAUAUAUAAAAUGA
AAAUCUCGCUGAGGAGCGGAGCACUACCACAGCAGAUUUUUCAAACCAGAACUGUGUUUA
UUCUCAACGAUAAUGGAAAUUACACCAAAAUGAGGGGACAGUCAACAGUUUUCCGGUCUC
CAUGCUAAUUCUUUGGACCUACCAGGUUUCCAGAAGAGACUUCUACUUGCUAAUUCUUUG
GACCAAACAGGGUUCCAGAAGAGACUUCAAUGUCAAUGGUUCCUCAACAGGUUUCUUUUA
AGUGUUUUGUGGUAGGACGCACACGGACAAGAAGGCGCCCACCCCUCAUCUUCUACAAGA
GAUCCCAAGCACUUGGAUUGUUUGAGCUUUUCGUUCAUGUCAUUAAAAAACCGGGAGAGU
AGAUAUGCCCCCGGUAAUCUGUCUUUCUUCGAUCUCUGGCAGACAAAAAGAAAUACACUA
AAUUAACUUUUGUAGCAAGCUGCAUGGACCGCAGCACAUAGCCAAAGGAGCUUGGAAUCG
AUGAUAAGAAGAAGGUAGGAAAAUAUAAAGGUUGAAAACUAUGGCUUUGCGAUUGUACCG
UAAUUAAGUAUCAGCACCAAACUCUGAAAAACAACUGUACUAAUUAAGAAUUUUAUCAAC
UUAGGUCAAACAAAGAAAACAUUGAGCAGUACAGAGAGGUGAAGAAGAAAAAGAAGAGCA
GGAAAGAUAAUCAGUGCAAAUACAAAAGGAAGGAAUAAGUCACAGAAAUGGAGCAGAAAG
CAAUAGAAAAAGGGGGAUAGUGACGCACCAUUGGGAACACCAUUCAAUUUUGAUAGGAAC
CCAUCUUUUACAAGGGAUCAAGACUAACACAAUCAAACAACACCUACAGUGACUUACAAG
UAGGAUGACAUACUUGAGGAACGAGAAUAGCAAGUUGGAGACAGUUGUUCCCUUGACAAA
AGGAUUGUAUGUAGACUUGUUAAAAUUUAUUAAACUUUAAAUCUGUCAGUCUUAUCAUUU
GGAAAAAUUCGAUGAAUUUAGUAGAUUGUUCUAGCAGAGCUUUUUUCUAUGCCUAAGCAU
AAAUGCUUGUUCUUAGUCAUCAGUCAUGGCUCUUGCUUAAUGGUUUGAUCUUGGACAAGG
AACUAUGGACACACUAGAGACUAAAAUUAUAUUGCUAAAAUUAAUUAGAUUGGUGGACCA
UAACCUCACUGGCUCACUAGAGACUCCCAAAAGCUACUCCGGUACUCCCUAUCAGCUACU
AGCUUCUGCAUUACAAGGGGAUGUGGAAAUAUACGAUUACAUUAUCGUUCUUUUUUAUAA
GAAGAAUUAUCAGAUAACAUCUUUAAAGCUGCAAAACUAAGGUCCAAAGUAUGAGAGCCG
AUUCGUGCAUGCAUAUUCUACAUUCUAGUUACCACAAUUAUAUCAAAAGAAUGUGUACAA
CAACUAUUACUACACAUUAGUCCCAAACAAGUUGGGGGAUCAAAAACUGUGUCACAUGAC
UAAAGAAAACUUGAGACUUGUAAAGCUACUCAUAUGACCCACAUGAUGAUAUGCCAGAAA
AUCCACUUUGGACCAAAGGAAUUUGUUAGGAUCUGAUAGUACAGGGUAAAGUACUAAUUG
UUCAGACACACUUCUUUCCUUAAAUUUCUGCCUAAUGCAAAAGAAAUACAUAGAAGGGAU
AUUUGUGUUUUUAGCUUUAGUUUGGCAAGAACCAGCUUCGAUUUCCAGCAAUACUUGGCU
AGACAUAUUUAAAUCCUAAAUACUGGUGAUAUCAUGAGCUAAUGCAAAAGAUUUUAUCAU
UAAACAAAAAAAGACCAAAAAAU
2. MFE structure-
..((..(((((((((..........((((((.(((..((...(((((...)))))...))..)))))))))..........))))))))).((((((((((((..(((((((((...........(((.(((((.....(((((...((((((((((((.....((.(((.(((((((((((((((....((.......)
).....))))))))...((((.(((((((.....)))))))..))))..)))))))......))).))..))))))))).))))))))((..(((........)))..))..((((((.......))))))....))))).)))))).)))))).))))).)))))))....((((((...(((........)))...))
)))).(((.((.(((....))))))))..(((((((((((((.....((..(((((((((................))))).))))..)).))))))......))))))).....((((((......)))))).(((((.((.....)).))))).........((.(((..(((...)))..))))))).(((((((..
........(((((.....(((.(((((((((...((((......)))).............((((.......)))).......((((....)))).........(((((((((((((((..(((.(((((((..........)))))))...(((.....)))...............((((((((.((((.((((....
...)).)).))))))))))))......((((((((......((((..((....(((........)))...))..))))......))))))))..(((((.(((((.(((((((((((...((...(((((((((((...)))))((((((((.(((((((((((......)).)))).....(((......)))....((
(((((((..(((.((((((...(((((((((........)))).))))))))))).))).........((((....))))((((((..(((((..((((((((..(((((((.(((...((((......((((((...(((....))).))))))..(((..((((((((.(((((..((..((((((............
.......)))))).))..(((((.......)))))......((((......))))....)).)))..))))))))..)))..........(((((....(((....(((((.......)))))....))))))))......((((.........(((((((..((((...((((....................))))..
.))))..)))))))))))))))....)))...))))))).....))))))))))))).......((((((.((.....(((((....................................)))))...))..((((....((..(((....)))..))..))))))))))....)))))).....................
(((((((.(((...)))..))))).))......)))))))))))))))))))))).)))))).))...))).......))))))))))))).)))))..................(((((((.((((((((.......(((((((...(((..........((((....))))...(((((...((((.((((.(((((.
...)))))((((((.((((((((.((((((....)))..)))..)))))....))).))))))(((((...((((((...(((..((((.(((.....)))))))))).))))))..)))))......((((....))))(((((....(((((((....((((((((((...((((...))))((((((((.......)
))))))).....(((((......)))))(((((((...)))))))((((((((((...............(((((.((..((((((((((......((((...)).)).((((((((((......(((((.....))))).......)))))))))).....((((...........))))(((((.....)))))((((
........((((((((((.............((((.(((...)))...)))).........)))).))))))........))))))))))))))..))...)))))....))))))))))..))))...))))))........(((((((.(((((((.....((((((............)))))).....))))))).
...))))))).))))))))))))))))))))..)))))))))))))))........)))).)))).)))))))..)))..)))))))))).)).))))))))))))...)))....)))))..........))))))).....
You can download the minimum free energy (MFE) structure in
II. Results for thermodynamic ensemble prediction
1. The free energy of the thermodynamic ensemble is [-637.95] kcal/mol.
2. The frequency of mfe structure in ensemble 1.65102e-34.
3. The ensemble diversity 575.91.
4. You may look at the dot plot containing the base pair probabilities [below]

III. Graphical output
1. A note on base-pairing probabilities:--
The structure below is colored by base-pairing probabilities. For unpaired regions the color denotes the probability of being unpaired. The reliability information of RNA secondary structure
plot in the form of color annotation, Positional entropy ranging from red (low entropy, well-defined) via green to blue and
violet (high entropy, ill-defined).
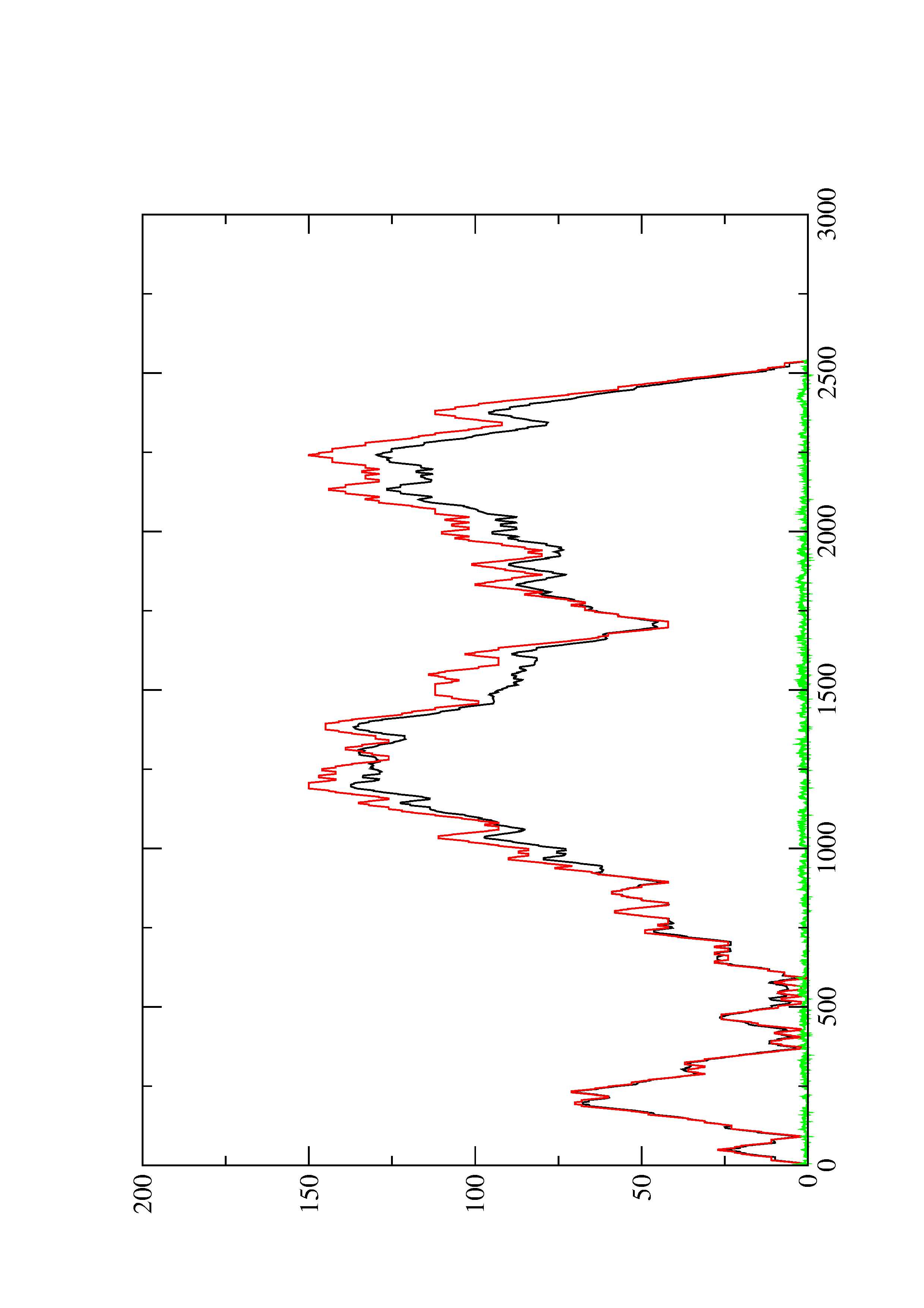
2. Here you find a mountain plot representation of the MFE structure, the thermodynamic ensemble of RNA structures, and the centroid structure. Additionally we present the positional entropy for each position. The resulting plot shows three curves, two mountain plots derived from the MFE structure (red) and the pairing probabilities (black) and a positional entropy curve (green). Well-defined regions are identified by low entropy.