lncRNA Structure Prediction
Species Name : Withania somnifera Sequence Id : GBHJ01027264.1
I. Result for minimum free energy prediction
The optimal secondary structure in dot-bracket notation with a minimum free energy of (-457.90) is given below.
1. Sequence-
GUCGACUUUCUUUUUCCUUAGCUUCUCUUUUUGUGCUGGUUUGGUUCAAAUAUGAUAGCU
GAUUAGGCCCCUGUUAUCGUCGUAGCAUACAAUGUCUGUUAAAUUGAAAUUUAUUUGUAG
UUCUUGUCUUACUUGUGAUUAACUAGAUUACUUUUGAAUUCUCUUGCAUGCUAUGUGCUA
AACAUAUUCUAAUAAUUUGUUGCACUUCUAUGCCUACGUAUUCCUAUUUUUCUCAAGUAA
AAGAUCAAAUUGGUUCUGUGUAACCUUCCUGUCUCCAGUUAGCUUAUAGAGUAUUAACAU
UAGUCUUAACUUGUUUCCUUGUUUCUGGAUUGGCUUAUUCCUAAUAUCUUAUUCAUGUGA
AGCUAAUCACUGUGUCUAGAUAUAAAUCCUAUUUUAUAUGACCUGCUCAGUGAACCUUUG
CAUUGAACAUAAUAUUUAGCAGUUGAGUUUUUUUAAGAAAAUUUGUAUCAUCUACUUGCU
UGUUUGAGUUUUUCUCUUAACUACUUUGUAUGGAAGGCCUGCUUAAAAUGUGAUAGCUAA
GAGUAGUUUCCACUAAGUCCUAAUUCUUCCUAAAAGAAUAUGAGCAUAUGAUGUCUGUUG
AAAUCAUUUGACUGUCUCUUGUUUCCUACCUUACUGUGUUUGUUCUGAAUUGUCUUUUAU
AGUCCCUUACUAGCUAUAUUAUUAGAGUUUUUUUUAUAUCUUCUCCACCCCCUUGUUGAC
CUAGACAAAGGUUUGCAUCAUGAUUCCUUUCCCUAAGUCCCAACAUGCUGUUUUGUUACA
UUUUUUUUCAUUUACAUUAGGUUAAGUGACUUCCCUGCUAGCUUGCUACCUAUUUAUUUU
CCUUCAUACGGAGUUCAUGAUAAACCUGCUACUUGUCGCUAUGCACUCUUAUUCACAAAG
UCUAUGUUUUUACUUGCUACCUCCCCCUCUCAUUCCUUUUAUACUGAAGCCUAUUGUAAA
CUUUCUUGCCUAUCUUCUUAGUCUCUCUUCGUAUUAAGUUCGCUCCAUUUUCUUAUGAAC
CUGCUGAUUGCCUUUCAUUUUUCCUGUCCUUUAUAAACUAAGUUUAGGUUGUAUGAUAUA
AACAUGUUAUAUGUUCUAAUUUCCUUCAUACUGAGUGGAUGCUGCCUUAUAUAAAUUGAU
GCUUUCUUUCUCUUUCUAUCAUAUUGAGUUUUCUGUAAGCAUACUAUUGUUUUUCCUCUU
CCUUAAUUUGUUGAAAUUAUGUUGUAACCUGUUGUCUUAUUAACUUGUUAAAGAUCAUUA
AUAGUGUUAUUCUCUGGUAUCUCUGUCUGUGAUUAACUAAUUUCAAGUGUCUCUUCAUGG
GAGUUAAAGUAGACUACUGAAUUUGCAUUAUAACAUGCCUAGAGUAGCUGUUCUGUGCUG
UAAGUGUUGAUAUGACAUGGUCAUCAUACCCUCCCUUGAUAUUGUCAUACGUGCUUGCUU
GUUUCUCUUUCUAAAACUAGUGUGUUGAAUCUACAUAUUGAUUAACACAUCUAUAUCUAU
AAUGUUCUGCUAUUCUUGUGCAUAUUUAUCCUCUGCUACAUUUGGUUUCAGCAUUUUGCG
UUAGAAAUGCAUGACCCCUUGCCUCAUUUCUUGCAUGCUACUUGCUCCUCUUCUCCAUUU
CUCUUAGCACUGAUUUUAGCAUAGCAUGUUUUCUUUGCAUUCUCGCUAUAUACACUGGUA
UAAUCUAAUCCUGAAUAGAUGAACAUGACAAUUAAAAAUAAAUAAUAGUCAAAGUAAUGU
GUUCUCUAUAUUUUGUGUACAUAUUGCUUGUCUACUUGUUUCGGAUAUGGUUUUUUAAUC
CAAACAACCCUAUUUUCUUGAAAGAAGUUGAUUAACAUGUAUCAUCUGCCUUCCUCUAAU
UUUAUUUCUUAAAUGGAUUGUCAUUAGAUUAAUUUCGAAUGCUAAAUAUUUAUACUUGUC
UUAUUUGGAUUCUUCAUCAUGUCUCUGUGUGCAUAUAGUAUGUAUCACUUAAGACUAUUA
AGGGGAUUAAUCAUUAUUAUACACGCUAAGUGGGUAUUGAACUAUUUAUUUGGUUAGUUA
UUGAGUUCGUGAAAUUGGGCCAGAUUGUGCAAGUCGAUUGGCCCAAGAUUUCUUUAAUUU
UAAUUCUAAUUAGUCUACCUUUAAUCUUCUUUAGCCCAAUAGUCAGCUGAUUUUACUUCU
UUUGGGCCAUUUAUUUUCUAUAUUUUCUAAACUAUGCGUAUUUGGUCCUAGAUUAACAUA
ACAUGAUGUUGUUAGAUUCAUCUUUAUUUUCCUUGUUUCAUUUAUUUACUUAUAUUAUAU
AUUACUAUCAAUUUCCUUUUUUCAUGGAUAAACUUGGUCCCUAGAUUUCCUUGCAUGUCC
UUGGGGGGUUAAAGGCAAACCAAGUUGAAGAUUGGACUCAGUCCGAAUAAGUUCAUUCAU
UUUCCAUGUUUCCC
2. MFE structure-
................((((((.......((((.(((.....))).))))((((((((.((((((..(((((...((.(((.(((((.((...)).)))))....((((((((...((((((...(((........)))......))))))...)))))).)).((((((((((((((...(((((.......(((.(((
.(((......)))..))).)))...........(((((..(((((....(((((.((.(((((.((..(((....)))..)).))))).)).))))).....))))).)))))..(((........)))..((......))..........(((((((((((....((((((.((....(((((((......))))))).
....)).))))))..))))))).))))...(((((((((((.((((((((((....))))))..........(((......(((.(((.....)))..)))......))).(((((((..(((((....(((..(((((....)))))..))))))))....((((((.....))))))..(((((...((((((...((
....))...))).)))...)))))........((((((((((((...(((((((...((((((.........))))))......(((...............))).......(((((......)))))((((((..(((((((((((..............((((........))))....................(((
((..(((((((........)).)))))))))).................)))).)))))))))))))(((((((...((.(((((.....(((((...(((((((.....................(((((((.......(((((((((......((((......))))....))))..(((.((..((.((((.(((..
.(((((((((...((((((...((.....))...((((....((((..(((........)))..))))....))))....((((((...)))))).........))))))..))))))).))...))))))).))..)).)))................((.((((((........(((((......)))))........
.)))))).)).((((((((.(((.((((..((((......))))..))))..((((((..((((...((((....))))...))))..))))))))))))))))))))))......))))))).....)))))))..))))).)))))........))...)))))))...........((((((..((.(((((((..(
(((.............))))..))))))))).))))))...................(((((((((((.((........)))))))))).)))........((((((..((((((.((((((...........))).)))...((..((((((((((......))))))).)))..))..............((((((((
..((((.....((..................))....)))))))))))).......((......))..(((((....)))))..........))))))..)))))))))))))...))))))).)))))..(((((((((((................)))))))))))(((.((((.(((((((((.((((.(((....
.))).)))).))))....))))).)))).)))...............)))))))((((((..((.(((......))).))..))))))......)))).))))))))))).......((((.....))))...............)))))))))))))))))))........))).))..)))))..)))))))))))))
)...))))))((((...((((..((((((((((......(((((((((..(((((((((((((.(((.......)))))))))))..))))).((((((........)))))).........(((((((.(((((..........))))).............((((((..(((.......................)))
..)))))).((((.((..(((((......)))))..)).))))..................................................(((.........)))..((((((((.(((......((((.((......)).))))....)))...))))))))))))))))))))))))))))))))))...)))).
..))))........
You can download the minimum free energy (MFE) structure in
II. Results for thermodynamic ensemble prediction
1. The free energy of the thermodynamic ensemble is [-500.21] kcal/mol.
2. The frequency of mfe structure in ensemble 1.53126e-30.
3. The ensemble diversity 598.13.
4. You may look at the dot plot containing the base pair probabilities [below]

III. Graphical output
1. A note on base-pairing probabilities:--
The structure below is colored by base-pairing probabilities. For unpaired regions the color denotes the probability of being unpaired. The reliability information of RNA secondary structure
plot in the form of color annotation, Positional entropy ranging from red (low entropy, well-defined) via green to blue and
violet (high entropy, ill-defined).
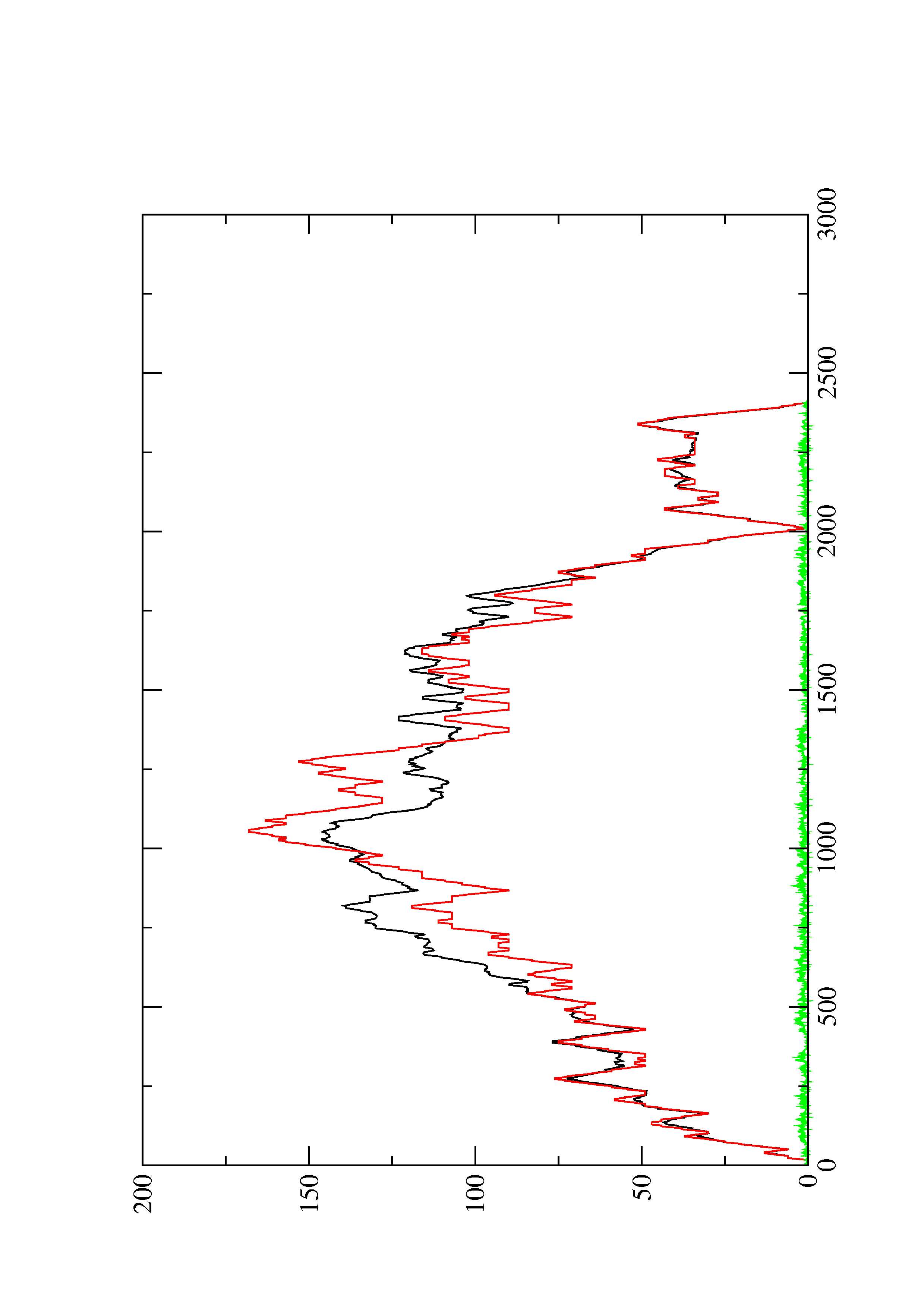
2. Here you find a mountain plot representation of the MFE structure, the thermodynamic ensemble of RNA structures, and the centroid structure. Additionally we present the positional entropy for each position. The resulting plot shows three curves, two mountain plots derived from the MFE structure (red) and the pairing probabilities (black) and a positional entropy curve (green). Well-defined regions are identified by low entropy.