lncRNA Structure Prediction
Species Name : Withania somnifera Sequence Id : GBHJ01027335.1
I. Result for minimum free energy prediction
The optimal secondary structure in dot-bracket notation with a minimum free energy of (-640.50) is given below.
1. Sequence-
CCAACUUAGGCUUUUUCACUAACACAAUUCACUUUCUAGUUCAGUGAGAACUCUUACAAC
GUUUAUUGAAUUUCGGCUCAAGAUACCCUUUACACUUCUUGAAUUUUGGUUCAAGAUACC
CUUUACUCUUGGAGUUGCACCCCAACCCCCUACAAUCACUUUCAAGGUAAGUGAGAACUC
UUACAUUAACCAAAUACAAAGAUUGAAAUCAAAUGUACAAAGAUUUGGAAUAGAAGAAUA
AGCUCAAGAACUUUGUACUUUUUCUUUGGUGAUGAACACUUGAUUUUUGGGUAGAAAUGA
UGAAUAAUGUUGUUGUGUUUUUCCCUUAGUGUUCCAUCAUCAGCAAAUACACUUCCAAGG
UGUAUUUAUAGCUCAGUUGGUUGAUAUAGCCGUUUGUGACCGUUUUAUAGCCGUUCUACU
GUUUUACCAACUGCAUUUAAUACUUCUGACAAGCAGAUUAGUCGACUACUUUGUGAUAUU
AGUCGACUAGUUUUUGUUCCAACUGGAUUAGUCGACUAGUUUAUACAAUUGGUCGACUAG
UUUGUCUUGCAUCCUUUUUGGUCGACUAGUUUGCAGUUUUGGUCGACUUGUUCAUUUUCC
AGUAUGCUACUGGAACUCAGAUUAGUCGACUAGUUUCAUAAACUGGUCGACUAGUUUCUU
GGAUUGGUCGACUACUUUGUUCAAUUGGUCGACUAGUUUCUUGGAUUGGUCGACUACUUU
GUUCAAUUGGUCGACUACUUUUGGAUUUCCAGUAUCCUACUGGAGCUUCAGAUUAGUCGA
CUAGUUUACCCAAUUAAUCGACUAGUUCCCUUCUUGUUCUUCACAAUUUCUUCUUUUGUU
UACUCGGAAAGGUCCAAGAUGAAUCCUUGACUUCCCGGAAUGUUAUGCUUGCUUGUGAAG
UGUCUCAUGGACAUUUUCUAUGAUGUGUAGGAGAUCCAUUAACAUCGAAUCUUUGACUUA
AUGGUUUGGUCAUCAAUCAAAACACGAAGGGACGAUUCUCCUGAACCAACCUUGGUCCAA
CACAAUUUUCUUGUAUGGUCCAGGCCUUAUGACCUGCUUUACCAUCUGAAUUUGUCCAUC
AGUUUGUUAUUCUCUUUAUCCCUAUUCAUUCUUUGUCAAACCUUGUUUCCAAGUUUUGUG
UUGAGAGUAAAAAAAUGAAUUAGAUGGGAAAGCUUAGAGUUUUCUUUGUCCCAGCUUGUG
CAUUAUGUGAUGAGCGUGGAAGAGUUCAGUAGUUACAUUACUUGGUUCUCUGAUUAGUUU
GAGCUUGCAUGAUCUGUAUAAGUGAAGUUUGAAUUUAGUGCUAUAAUCAUGAGAACAAAG
CAUCAUAACAAUUGGUUUCCAGUAGGGUGCAAGGGCCAUGGUGUGUUUAAAUAUCGAUAC
UCGAUAGACUGAUUUGAAUUUGCUACUUCCUAGGUUUGUUGGGAUGUGCCUUAUGUUCGA
AGAUUCAUUCAUGUCUCGAUUUCCCUGUGUGCUCCUUUGGUCUCUUUUUCUAACCAGAAG
CCACCUAAGUUAUUAUAUUCAAAUCUGCUUUGGUUAGUUGUUUAUUGUCAGAGAUUUCAU
GGGUUCUUCAUGGCGAUUAAAGUACAUAAUCUAUGUCUUGAGGCUGCCUUUCUAGCUGCU
ACCAGGACAUGAGCUCAGGAUAGAAAGAAGCCUUCCCAAUGCAUCAUUUCAAUCUCUGCA
UUUAUCACCAUUAUAGUACAAGCUGGAAAAUAAAUAUUUGCAUUGUGAUUGCAAGAGUUU
UUCUUACAUCUUCAAUAAGGUUCUUUUUUUGUUAUUGUUUUGUUUGGUUUAUUCAUAAAC
UUGUUUCCUGCUUUCAUUAUGAUUACCCUCCUACGAAGCAUGAGUUUCUGCUUUGGAUUA
AGUACAGAUUUCUAUCUUUUUUCCCUUGUAAGUAUUGUCGCCUGGAUAUGUCUACAUGGU
CGAAGCAUGCAUGUCACUAGAUUAAGAGACAUAUUGCUAGUUGAUUUUUGUUUGAAUUAU
UUCGAGAAUUCGAUCGAAUGCUAAUUUUAUUUUUCUUUUCUUUGCAUGAUACCUUCCACC
CGAGUCCAAUUGGUCUCUAUUUUGUUUAUCUUCUGAUGGGCCAGGCCCAUUUCAUCGAGU
CAAACCUCCAAGAUUAAAGUUCAAACAAGAAGUAAAACAAAAUGGCGUAUGAGCCAUACA
CAAUAAAGGAAGCUAAUAUACGGGGAAGACCUAAGCGGAGCUGAAAGAGAAAAAGUUUGG
GUAGUCCAUUUAGGACAUUCGGUAGGGGUCUGGGCCUAUUGGUCUUAGUAUAACUUCUCC
UUAUCUUGUUUUUAUUUAUUUAGAAUUGAUUUGUCUAAUUUCUAACUUUGUUUUCAACUU
GGUUCUACUUUUCCUUUGGAGUAACAAAA
2. MFE structure-
......((((..(((((((((((...............))).))))))))..))))....((...(((((((.....((((((.............)))))).....)))))))...))...(((((((.(((((((.....))))................(((((((((.(((((....(((....((((((((((((
(((((....(((((........))))).....((((((.(((..(((.....)))..))).))))))(((((((((((((........(((.(((((((((((......)))))..)))))))))..)))))).)))))))(((((((((((((...))))))))))...)))((((((((.(((((((.((.((((((.
....)))))).))..))).))))..)))))))).............((.(((((((((((((((((((..(((((..((((((((((((((((..((....)).))))))))))))))))...))))).))))))))))))))).)))))).((...((((((((((((((.((((...(((((((((.(((((..((((
((((...))))))))...((((((((((((((((((...))))))))))))))))))..))))).)))))))))..))))..))))))))))))))......))(((((((((.((..(((...(((((((((((((..((((((.((((((((...)))))))).)))))).)))))))))))))....))).)).)))
))))))(((((((.((((((((((((..........(((((...(((..(((((.(((.......)))))))).))))))))..........)))))))).(((((.((.(((((......))))).)).)))))(((((((..((((....)))).))))))).(((((.....))))))))).)))))))........
.((.((((....)))).))...))))))..))))))((((((((((......((((((.((((.......(((.((..(((((((.............((((.((((((((((...((((.((((....)))).))))....))))........)))))).....))))..((((((.(((((((((((.((((((((((
(((.((((((((((.(((((.((((((((((((...))))))..((((((((((((((((((((((.(((...........))))))))))))).........)))))).))))))...(((((......(((((...)))))..)))))....(((((((.(((((........)))))((((..((((((..((((((
(((.((.((((((.....)))))).))))...........))))))).))).)))))))....((((((.....((((((.........(((((((((.((......(((......))).......)))))))))))..........))))))....))))))....)))))))(((((.......))))).((((((((
.(((.((.......)).)))..))))))))))))))..(((.((((((.((.........)).)).)))).)))))))).))))))).))).)).))))))))))).....((((.((((((.......)))))).)))).................((.(((((((.((((...((((((((..(((....((((((..
..((.....)).....))))))..))).....)))))))).((((((.(((((......))))).))))))...............(((.((...((((.....))))...))))).(((.((((....((((.((..(((..((((((.(((....(((((((((....((((((....)))))).....)))))...)
)))....))).)))))).))).))))))....)))).))).(((.((....)).)))...(((((((..((((((((((((...))))))).....(((......)))...................)))))..)))))))(((((......)))))..)))).))))))).)).....))))))))))))))))).)))
))))..)).))).......))))((((.....))))....))))))))))))))))..)))))....)))....))))))))))))))............(((((((((.(((.....................))).)))))))))....)))...))))))).....
You can download the minimum free energy (MFE) structure in
II. Results for thermodynamic ensemble prediction
1. The free energy of the thermodynamic ensemble is [-681.61] kcal/mol.
2. The frequency of mfe structure in ensemble 1.07213e-29.
3. The ensemble diversity 636.70.
4. You may look at the dot plot containing the base pair probabilities [below]

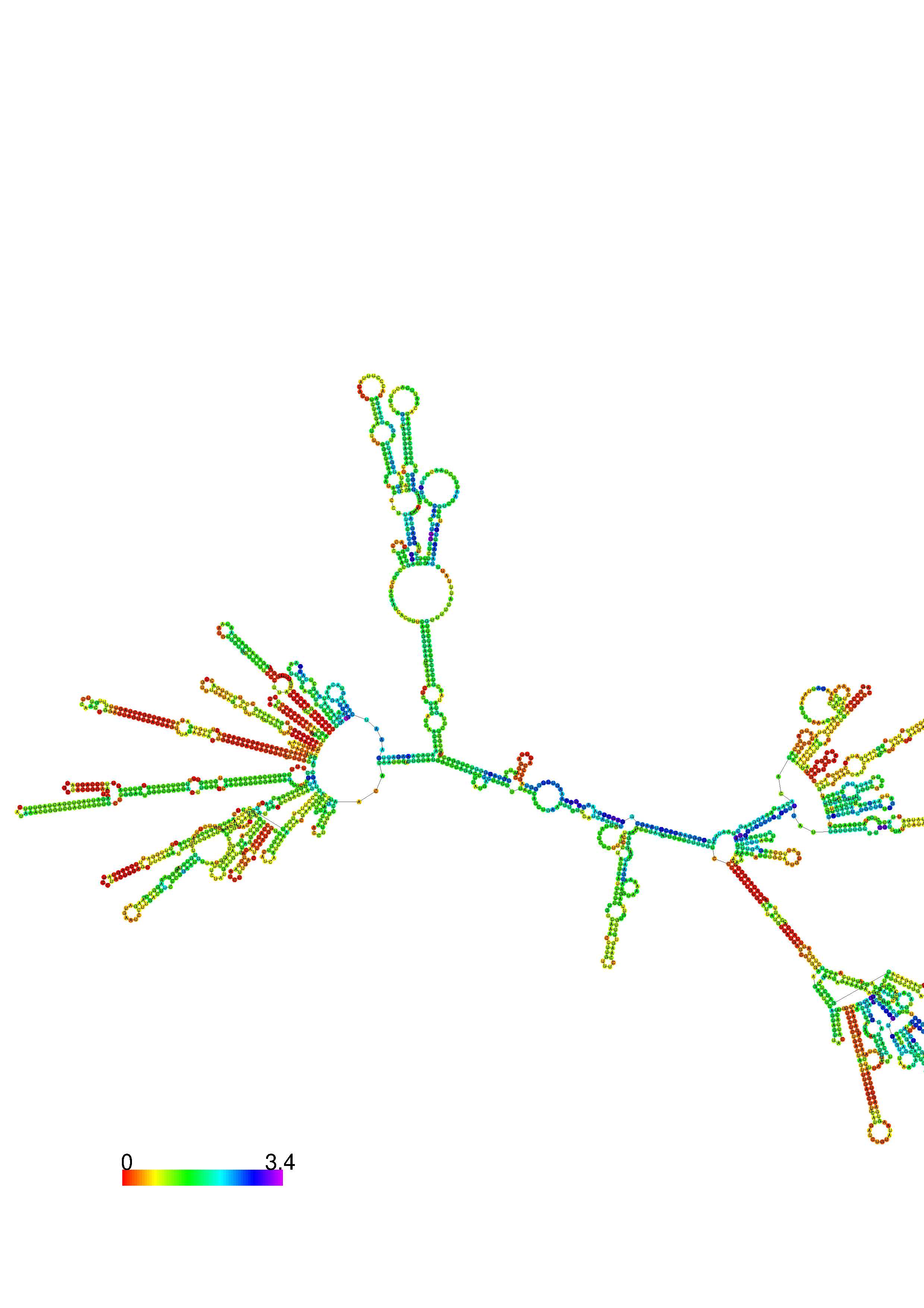
III. Graphical output
1. A note on base-pairing probabilities:--
The structure below is colored by base-pairing probabilities. For unpaired regions the color denotes the probability of being unpaired. The reliability information of RNA secondary structure
plot in the form of color annotation, Positional entropy ranging from red (low entropy, well-defined) via green to blue and
violet (high entropy, ill-defined).
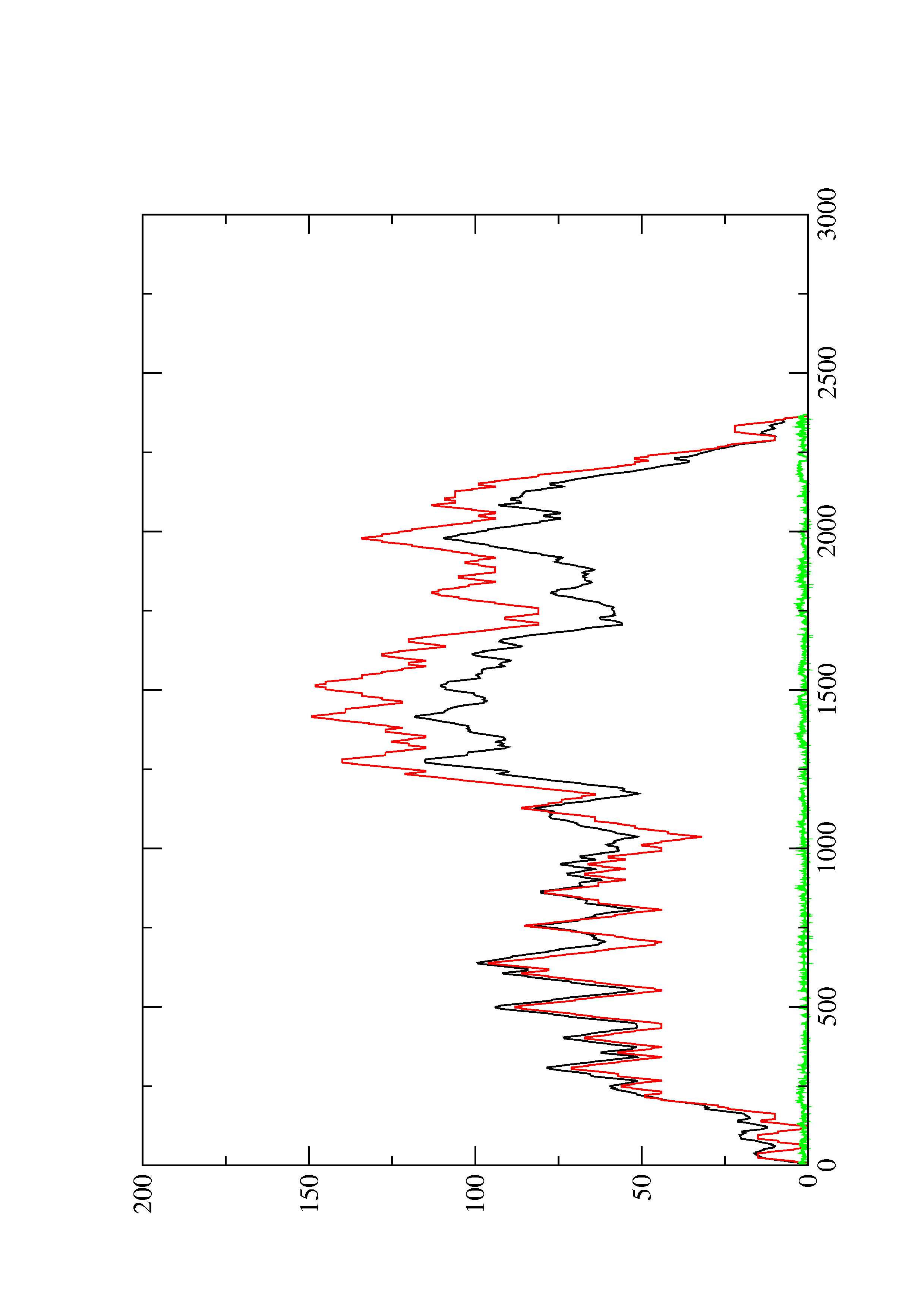
2. Here you find a mountain plot representation of the MFE structure, the thermodynamic ensemble of RNA structures, and the centroid structure. Additionally we present the positional entropy for each position. The resulting plot shows three curves, two mountain plots derived from the MFE structure (red) and the pairing probabilities (black) and a positional entropy curve (green). Well-defined regions are identified by low entropy.