lncRNA Structure Prediction
Species Name : Withania somnifera Sequence Id : GBHJ01028047.1
I. Result for minimum free energy prediction
The optimal secondary structure in dot-bracket notation with a minimum free energy of (-515.90) is given below.
1. Sequence-
CAUGCUAAUGUCUAAGUCAUUUAAAGGGAUGUAUAGGUUUCUGUUUGUCUAGUUCUUUGU
AUGUGUGAUUCCUGCCAUAUGUUUGAAAUGUAAGUCUAUACCAACAUCACGAGUCUUAUC
CUGUUCGAAAUGGUUGGUCUUGGAUGUUGACUCUGCACAUCUAGCCUACAAGACCUCUUU
GUGUCCUGUUUCACAUGUUUAACUGAGCUAGGGUGAGUGUCUUUGAAGCAGUAUGCUCUA
GCUGGUACCCUUGGCUUUAGUUGAGCUUAUUUAGUUUAACUGUUGCACCUGUCAGCCCAA
CUCUAGAUUUUCUUGAAAAUUAUGUGAAAACAUGCUAUGUUUGUUAGCAUAAACAGUUGU
CUAUAGUUUUAGUAUUGGUCUUGCUUGGUGAAAACUUUCUCUGAGAUGCAUUAACCUGGU
AGCAUUUGUUAUUGAAUCAUGAUAUAUAGUAGGAUUAGUUCUAACCAUGCAUUAAGAAUU
CAUUUGAUAAAAAGGUUUAUUCCCUUAUUUCAUAGUCUUAAAUGAUGCCUAUCUGUUUGA
UAAUCUGUUUGAAACAACCUGAUGAUUCUGAGUGCCAUACAUGUUUUGUGUUGUGAGUUG
UUAGGUCAGUUCUUAAACUCGAAUUUUUGAUAUUCGAAAGCCACAGCUUGGGGUACAUAG
UUUAAGGAUAUAACCCUUAGAUGUAAAGUAUUUGUAAGUCUAUGCUUGUUUCCCCUGUUA
UCGCUGUUUACCGGAAAAUGAAAGUUACCUUGAUCACUUGGUUUUGUGAUUAUCCAUGUU
GUCUUUGGAAGUGUAGAAAUGGGUCUUUGACAAGCCGAGAUGUUGUAUGGAAUUUUUUUU
UUUUUUUUUAAGUUGAGAACUUCUGUGGCUACUUUUCUGGAUGAUUAAAGCUGAACCUUU
GAUGAUUUCUCCAGUUUAAACUUUGUUUUUUCCCUUUUCUUUGUUAAUUUCUAUGUCGCA
GUUCUAGAAGCCUAUUAGUUCCCUUAUCUCUACUUGGUUUCAUGUGACUGUCUAAAAUCC
CAAUUUGUAAUCUUUGAAAUAUAUCAUGUGGUUAUCUAGAGUAUGCAUCAAUUUUCCCUA
GUAUCUUCUCCCCUAAGGAGAAUACAGUUAGAUGUUAAAGAGUUCUAUUAUUUUUGUUUA
CUUGAUAAAAAAUAGUAAAGGCCGCAAGGCUAUUACUAGUUGGGGAUAGCCAAUCUCUAG
AAUUAGAGGUGUGCAUGGGUCAGCCUUUGUUAUGAAAAUGCUUAAGUAUGUUUCCCUGUU
UCAGUUUGCUUUUCAAGUCAAUAUGAGCGUAUUUGAUUAUUUCAUGUUGUAAAAAGAUGG
GGUGUUUGACUUAGGAGCCAAAGAGGGUUGGUUUUUCUUUUUGAGGUAAUAGUUUCAUAU
UCUCAAAGGUGGCAUUUGGUAAGCCUAGAAGGUUGAAUGGUUUUGCCAAUAAACUGCUGU
CUGUUAGAGGUUAAAAGGUUCUGAUCUUUGGUUUAGUUGACAAUGAUAUUGGGAUUUCCU
UGCUUUAUUUUUCGUGUUGGAUGAGUUGUUUUGAAGUCGUUUGAUUGGUCAUUUAUGGUU
GAUCUAUAUAUGAGUAUGAAGUUUGGAGUUUGAUUUCUUGAUUUAAUACCUCCCUUUGAU
AGGCUUGAAGAAAUUGGUAAUAGAAAUCUUAGCAAAUUUCAUAGGGGGUUUGAAGAGACU
UCUUGAAUUUGAACCUUCUUUUGAGCUGAAGGAGGAGGAAAAAAACAGAUUUUAUUGGUU
AGGAGAAAAAUGUUUCUAAGCUGAUUGAAGAAAUGGAUGGAGAAGUUAGUUCUUGGAUAU
UUCUCAUACAGAUUGUCAUAGGGUAGAUCUGCAAGAUUGGCCUCUAGUCCUAUUAGUAGU
AUUCGGCCAGGAAGAAAUGAAGAUGCAAAAACUAGACUGGAAGAGGUUCUAUUUUACCCA
AGACCUAGCCAAUCUAUUUCUUGUCUGUUCCAGUUUACUUGUUGAAAAUGGCAAAUUGUG
CUCUAGCGUGGUAAUAGGUUAAAGAAAUAGGCCAGUAAUGGAUUAUCUCCAAUGUAUCAC
CAUCCAAACCUUUGUUGUGGUGGAUAUUUGAAAUCUGGCAAGGCUCAUAAGGUAUCCCUC
CUGUAACUAGUCAUAUCAGAAUAUCUUUUUCUUUUUUUUUUUUUUUUUUUUUUUU
2. MFE structure-
......................(((((((.....(((((((((..((.((((((.........(((((..(((((((....((((((..(((((.((..............)).)))))....)))))).((((((((((((((((.((......)))))...........(((((((((..(((..((((..(((.(((
((((.(((((((((...(((.......)))....))))))))))))...(((((((..(((((((((.....)))))))))(((.((((.((.((.((((..(((((...........((((((.((.(((((((((.......)))))).....))).)))))))))))))..)))).)).))..))))..))).....
....(((.((.((((.((....))...)))).)).)))..(((((.((((((((((((..((((...((((((((((.......(((....((((.......))))...)))...)))))).......((((((...(((((((((((.((((((.((((((((((((.....((.(((((.....))))).))))))))
))))))..))).......((((((.......)))))).((((((((...((((.((...((((((((.......))))))))...((((((..........))))))))..)))).(((.(((((.......((((((.(((((((.((.....((((((((((.((((..((((((.....................))
))))..)).)))))))))).))......))))))))).))))))......)).))).)))..))))))))......((((((.(((((((......))))))).))....)))).......((((((((((.((((((((((((((((((((((....(((((((..(((((((((((.................((((.
.....))))..........((((((......((((((((((..((((.((..(((..((((((.(((.......((((.((((((((((((.....)))))).(((((((((...........))))...((((((((.....)))))))).((((((.((((....)))).))))))(((((.((.((((((((((((.
...))))))).........((((((((((((....((((((((((((......((((((((...(((((..........((((((((.(((......)))))))))))))))).))))))))......)))))((((((((......))))))))..(((((((((..(((......)))))))))))).)))))))...
.)))...))))))))).))))))).))))).....)))))....(((((((((........)))))))))(((.....)))...))))))))))......)))..))))))..))).)).))))..))))))))))......))))))((((.(((((.....))))).))))((((.(((((.((((......)))).)
)))).))))......)))))))))))..)).)))))...))))))))..))))....((((.(((((((((((((((....)))...))))))))))))..))))...)))))))))).))))))))))(((.(((((((.(((((......))))).))))))).)))((((((....(((((.....)))))...)))
)))))).)))))))))))..))))))...))))...))))...))))))))))))...))))).)))))))......)))).)))...))))..)))..))))))))).........)))))))).))))).((((((((((.((((((((((((((((((((......)))))...)))....))).))).))))))..
.))))))))))(((....)))......((.(((((.(((((((.(((....)))..)))))))))))).))....)))).))).)))))(((.......)))..)))))).))...))))).))))...)))))))...................
You can download the minimum free energy (MFE) structure in
II. Results for thermodynamic ensemble prediction
1. The free energy of the thermodynamic ensemble is [-563.29] kcal/mol.
2. The frequency of mfe structure in ensemble 4.04723e-34.
3. The ensemble diversity 645.76.
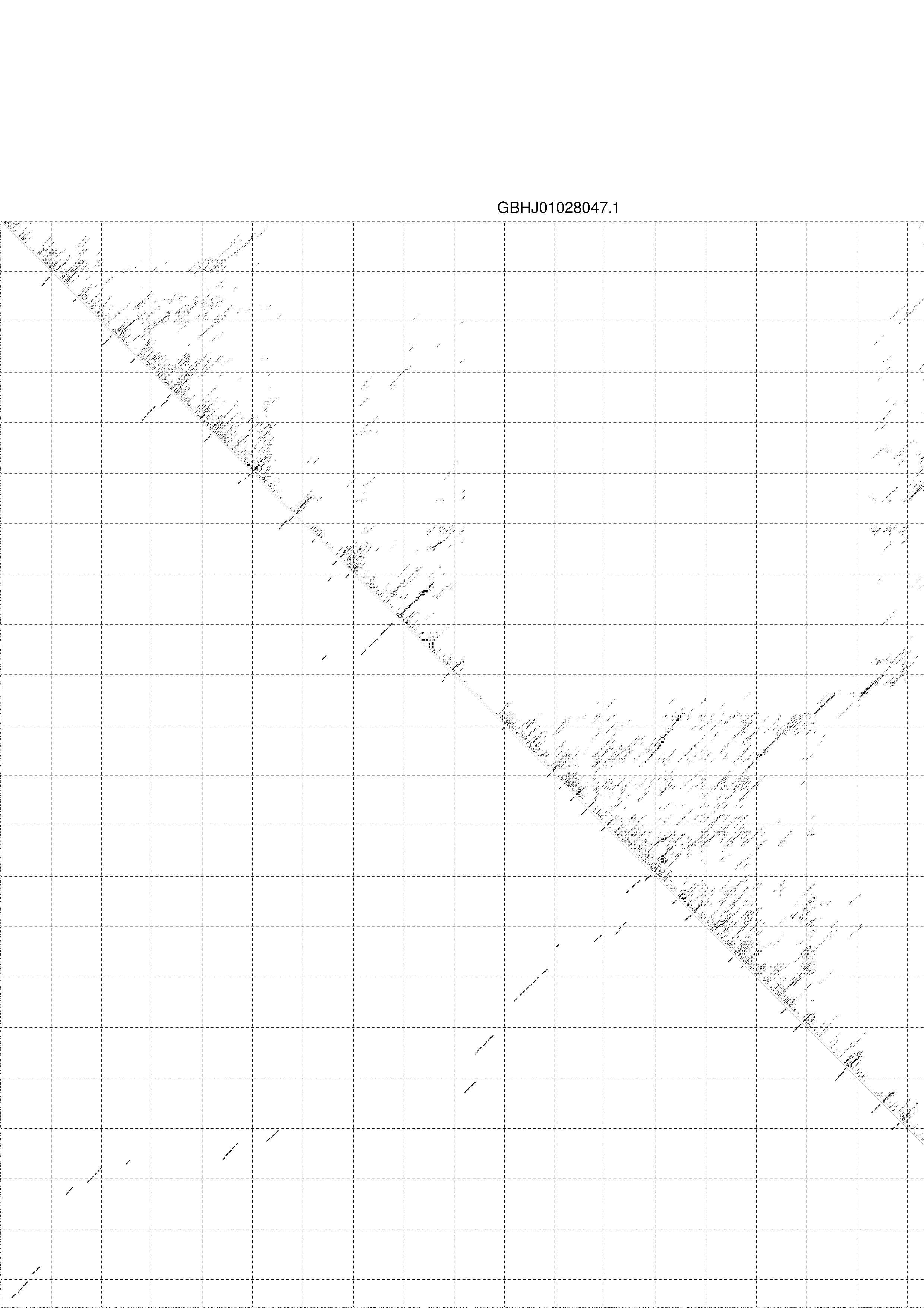
4. You may look at the dot plot containing the base pair probabilities [below]
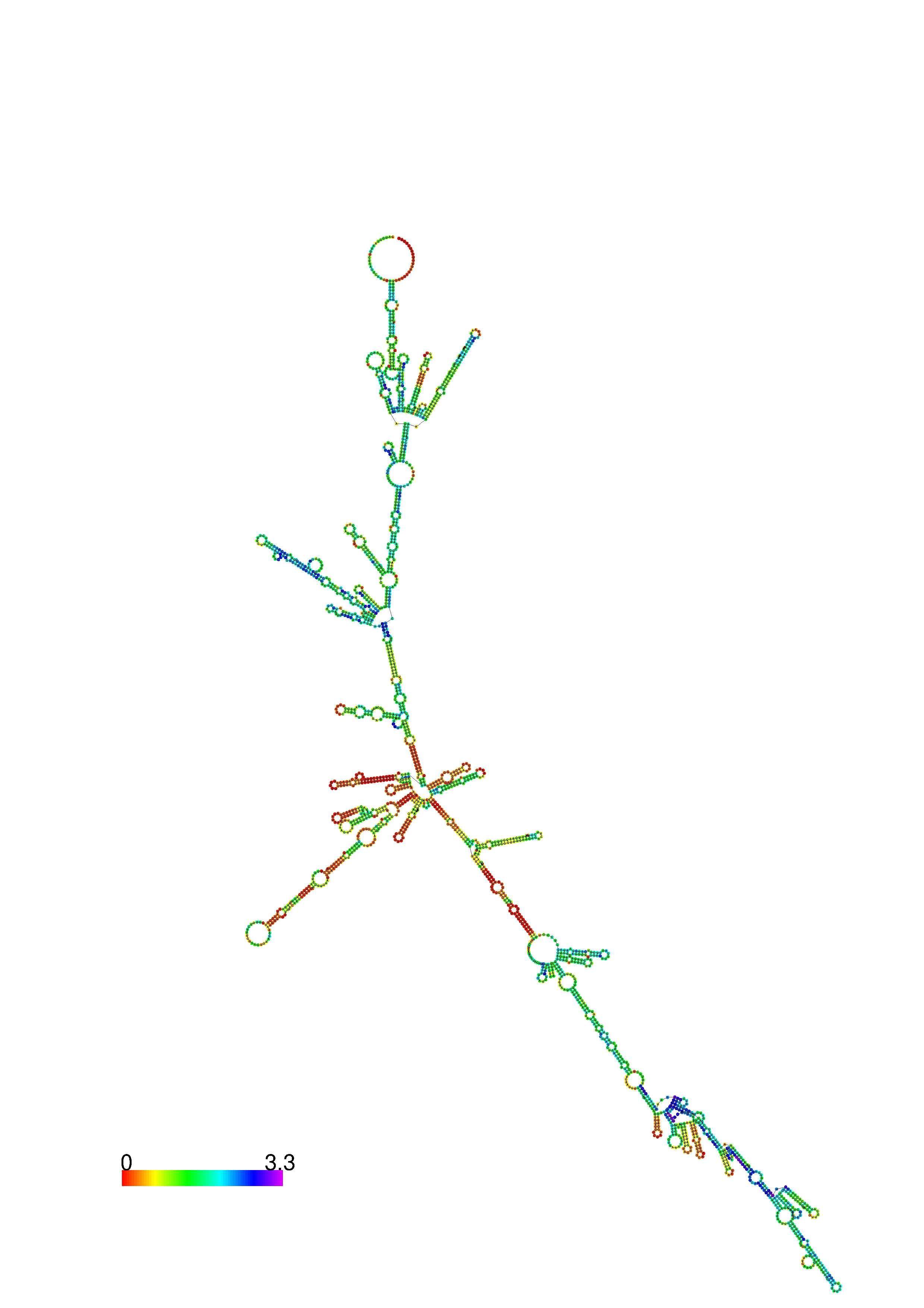
III. Graphical output
1. A note on base-pairing probabilities:--
The structure below is colored by base-pairing probabilities. For unpaired regions the color denotes the probability of being unpaired. The reliability information of RNA secondary structure
plot in the form of color annotation, Positional entropy ranging from red (low entropy, well-defined) via green to blue and
violet (high entropy, ill-defined).
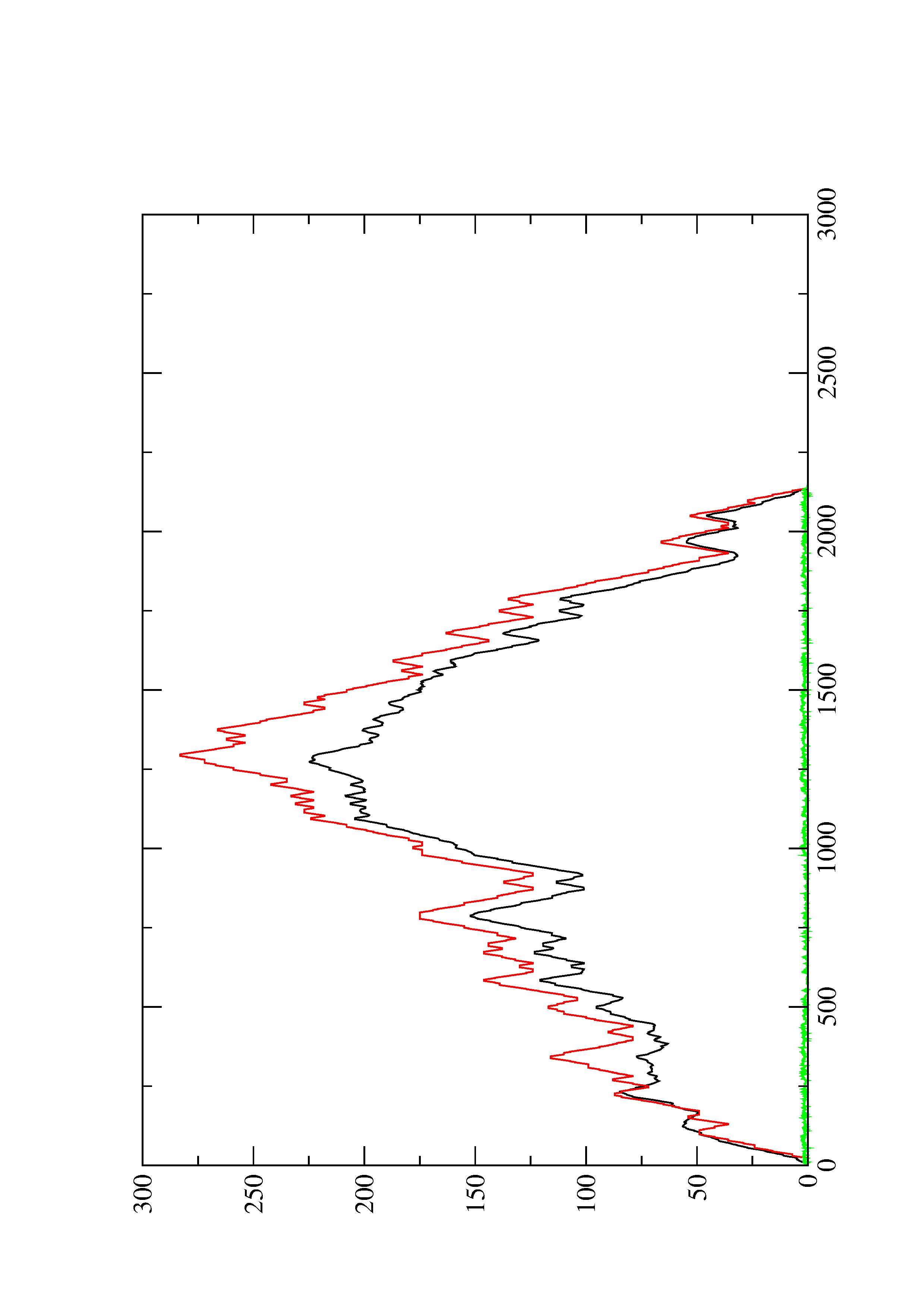
2. Here you find a mountain plot representation of the MFE structure, the thermodynamic ensemble of RNA structures, and the centroid structure. Additionally we present the positional entropy for each position. The resulting plot shows three curves, two mountain plots derived from the MFE structure (red) and the pairing probabilities (black) and a positional entropy curve (green). Well-defined regions are identified by low entropy.