lncRNA Structure Prediction
Species Name : Withania somnifera Sequence Id : GBHJ01029832.1
I. Result for minimum free energy prediction
The optimal secondary structure in dot-bracket notation with a minimum free energy of (-555.17) is given below.
1. Sequence-
CCGACUAUUGAACCACCCAAAAACUAGUGUCACUAUGUACAAAUCUCUAAACAUCAGGAG
UACAUAAAUGAAAAGAGAUACAUAUGUAUCCAAUGUCUAGGAUCCUCAAGUAGGAUCAUA
UACAUAAAUAAAAGUUUGGGAAUCUGUCGGAUAGCAAGCAACUACCUCUAGGACUAUCGA
AAUAUGCCUCAAAUUAUGAUCUGUGAAAAAGCAAAUCAAGCUACCUUGGGCUUUGAACCU
ACACACAUGUAGAAAAUCAAGGGGGUGAAUACCAAAAAUAAUCAAUGGAUACUUAAACAA
GUAUCCCUGGUUAGCCCUCAAACAAAGUCAUGAACAACAUCACUACCCCGAUGUACCGAC
CGCAAUCCUCCUAUAACACCAGGCUCUAAAAAAUAUCAUAACCAACAGUUUAUAUAAAAA
AAUACAGGGUGUAGGACAACUAUAGAUUAACCAAAUACUCAACCGAACCAACAAUAACAA
UUAUCAAAAGACAAAUAAUGAAGCAACGGAACCAAGUCUCAUAGACUAUCAUGGAAAUAC
AAUAUCAAUACUGCUUCAAAAGGGCAUCUCACCAGACAGAUAAUAACAUUACCAGUAUAA
AUUAAAUCAAAAAGCCACAAGUAUAUAAAACACACCCUCGUCCUAUUCCAAGUGCAGGAC
UCCUAAAUUGAAUUAACUCAUAACUCAUGGAAAAAUCCUACUAGUUGUCCAAUAGACUCG
CAAAGGGAGUCCACCAUCACUUAUAUUAGUCCCCAAUAUAUGAUCCACCUACGGUAGAAA
GGGUAUAUCCGAUGCGAUCGAGUAACAGAUCUAUGGAUAGGAUUCUAAAAGGAGUAGGCU
AAGAUAAAGAAAUCUACUCAAGUUUAUAUCCAUAUCAGUCCCUAAUACAUGAGUCCACCC
GCGAUAGCGACGGUAUAUCCAAUGAUGCCGAUAAAUGGACUCGUGGGGAAGAUUAUGAAG
UGGUUAGACCAAUACUAAUAUCGUAAUUCAUAAUCAAUGCAUAAUCAUCUCAUAACCUGA
ACAUAACCAUUUCAUAAACAAUGAAUACUCAUAUCAUAACCUGCACAUAAUCAUCUUAUA
CUAAUACAUGAUGAAUCUCAUUAUCAAAGAAUAAUCAUCAAGUAUAACCAAUCUGUACCU
UCCUUUAACACUCCACUCAGUCCCCGACUUACAAGUCCAUCCGCGAUACGAAGGAUAUAU
CCGACGAUACCGAGAAAUGGACUUGUGGGAUAGAUUCCUAAUGGGUAAGCCAAGAUACCA
UAAUGAUCGAGUGAACUGAUCUGCCAUCAUGAAAUAACUAUCAUCAGUCUAUCCGAAUAU
AGGUCCUCCUUAUCGAGUUAAUCCAUGAUUCAUGGGAUAUCCCACUCGUUAUCUACUAAC
UCGGGAAAGAGUUCUAUACACCCAUAUCAGUUCCCAACACACAAGUUUACCCGCGAUAGG
AAGGGUACAUCCGACGCUGCUGUGAAAUAGACUUGUGGAAUGGGUACUGAAGGAAUAACC
GUAAAAAUAUGUAUCCACACCAUCAAAUACAUAUAAAGAAGGAACUAGAAAUAGUGGGAG
UAUAUAUAUAUAUAUAUGAAAAAUCGCACAUCGACCCUGGCCUUUUAAAUCAUAUGUCAC
UAUCAUGACCCUGCCGAAAAUCACACCUCAGCCUUGGCAUGUUUGCAUUAUAAUCGAAAU
CAUAAAAAUACCAUAAUUUGCACCUCGAUCCUUGCCCUUUUUGUAUCAUAACCAUCAAUG
UAGUGAAAUAAUGCAUAAUGGACUUACAAAGCCACUUGUGGGCAUACUAGUACAAUCAUA
GACACUCAAGGGCAUAAUAUCAAGUAAAGAAUCAUAUCAAUCCAGCCAUGGCCACUCAAG
GGAAUCAUAUCAAGCAGGGAAUCAUGGCAUUCAAAUCAAAUCCACUCAAGGGCAUAUGAC
UUAAUAUAAGAACAAAUAAUGGCAAGUAAGUCGACGAGGCUCCCGGAUGGGCAAAUGAUC
AAAUACCAUAUCACCAUAAUGCAAUCAAGGCCUCUCAUGGGCACGCUUUCAAGUAAGGAA
UUGAUGAAAUAUAAUGUAACAUAUCAUAAGCAUAUCAAGGACAUAUAGAUUCUCAUUCAA
GUAGAAAUUAAAUCACCAAAUAUUUACUCGCUUUUAUUCCUUAAUUUUAACCCUCCAUGC
UUGGCUUUAGUGCCCGGAUGAGUACUUGUCAUUUCACCCACACGGGGAACUCCGAUAAUA
CCCAUUACUCCCAAUAUCAAGUAAUUACACAUAACAAGAAUAUUAUAACACUUGUAAUAU
GGUAUAACCUCAACUUACCUUAAGGAACGAGGAAGUAACUAAUCCGUGCCCUGCCUUUGC
GAUGUCCUAUCAAAUCCGCAAAAUCUAGUAAAAGAACGAAUUUUUCAGGAGAACUUAUGA
UAUGCAAUACCUUUAAAGGAAACCAUAACUUGGGUCCAAAUAUAUUCCCAAGAAUCACUC
CAAGAACCCUCUUAAAAAUCAAGAAGUUUUCAUGAAACUCAAGGCUCUGAUAGGUUGGGG
AUUUAAGUUAUUGAGUUAAAAUCAUAAGUUAGGGUUCAACUUGAUGGUUAAAAUAUCAUU
UUAUCCAUAAAAACAUGGGUCUUUAACCCCAAGAAAAU
2. MFE structure-
(((((.(((......((((((........(((.(((((((.....(((........))))))))))..))).....(((((....)))))..((((.((.((((((.....)))))).)).)))).........)))))))))..)))))..................((((((..(((((....(((.(((..((((((
.(((......))).))))...))..)))))))))))..(((((....)))))........(((((((...................((((((((....))))))))(((((..(((((........(((.(((...(((((........)))))..............((((...(((((............((((...(
((.....))).))))............)))))))))...............................................)))...)))......(((((((.........(((((...)))))....((......))..........)))))))..))))).....))))).........................
................................)))))))))))))...(((((...((((.(((((..(((.(((((((((((((...(((....)))....))))))......(((((.......))))).......(((((....((((((((((.....(((((((...))).....)))).(((((.(((.((((.
.......))))))))))))((((........(((((((..............)))))))........)))).((((((..(((....((((((((((..(((....))).((((((........)))))).....))))))))))((((.(((((((...((((((......(((.......))).....))))))...)
)))))).))))...................(((((.....)))))....((((((((((((((..........((((.(((((((((.((((((...((.........))....)))))).)))).((..((..((..(((((((..............(((((......((((((((((...((.((((..(((....)
))..)).)).))....)))))))))))))))...(((((...(((((((...((((((((((....(((.(((.((((((......................)))))).))).))).((((((..(((....((((((((....(((((...((((....))))...)))))....))))))))....))).))))))..
.((...(((((.((((...((((((((((..((((.(((....((.....))....))).))))...))))))))))...)))).))))).)).....((......((((((((............))))))))......)).((((....))))((((((((((((....))))))).....(((.....)))...((.
.(((((((.(((((((((.((.((((((((((((.((........(((..(((.((((.....((((...............................)))).((.(((...(((((((((((((......((((..(((((.........))))).))))...))))))(((.......)))...(((.........))
)......)))))))....))).))....................)))).)))......)))........))..))))))..)))))).....................))))))))))).))))...)))..))..((((((...(((.((((..(((..(((((..((((.((((((......................
.....((((..(((((.(((((((....(((((((.(((.....((((((...((((............))))...(((((...(((((..........((((((.(((.........))).))))))....)))))))))).))))))....))).))))))).....))))))))).)))..))))))))))..))))
..)))))..)))))))..)))))))))))))).....................(((((............)))))..))))))))...))...)))))))..))))).)))))))..))..))..))........((((((((..........)).))))))...))))).))))..........)))).....))))))
)))).....(((....))).........((((((............)))))).........((((....))))...........((((((...))))))..)))..)))))).)))))))))))))))....)))))))..)))....))))))))).)))))((((.((((((...)))))).)))).......((((.
.......)))).......
You can download the minimum free energy (MFE) structure in
II. Results for thermodynamic ensemble prediction
1. The free energy of the thermodynamic ensemble is [-600.56] kcal/mol.
2. The frequency of mfe structure in ensemble 1.02882e-32.
3. The ensemble diversity 446.19.
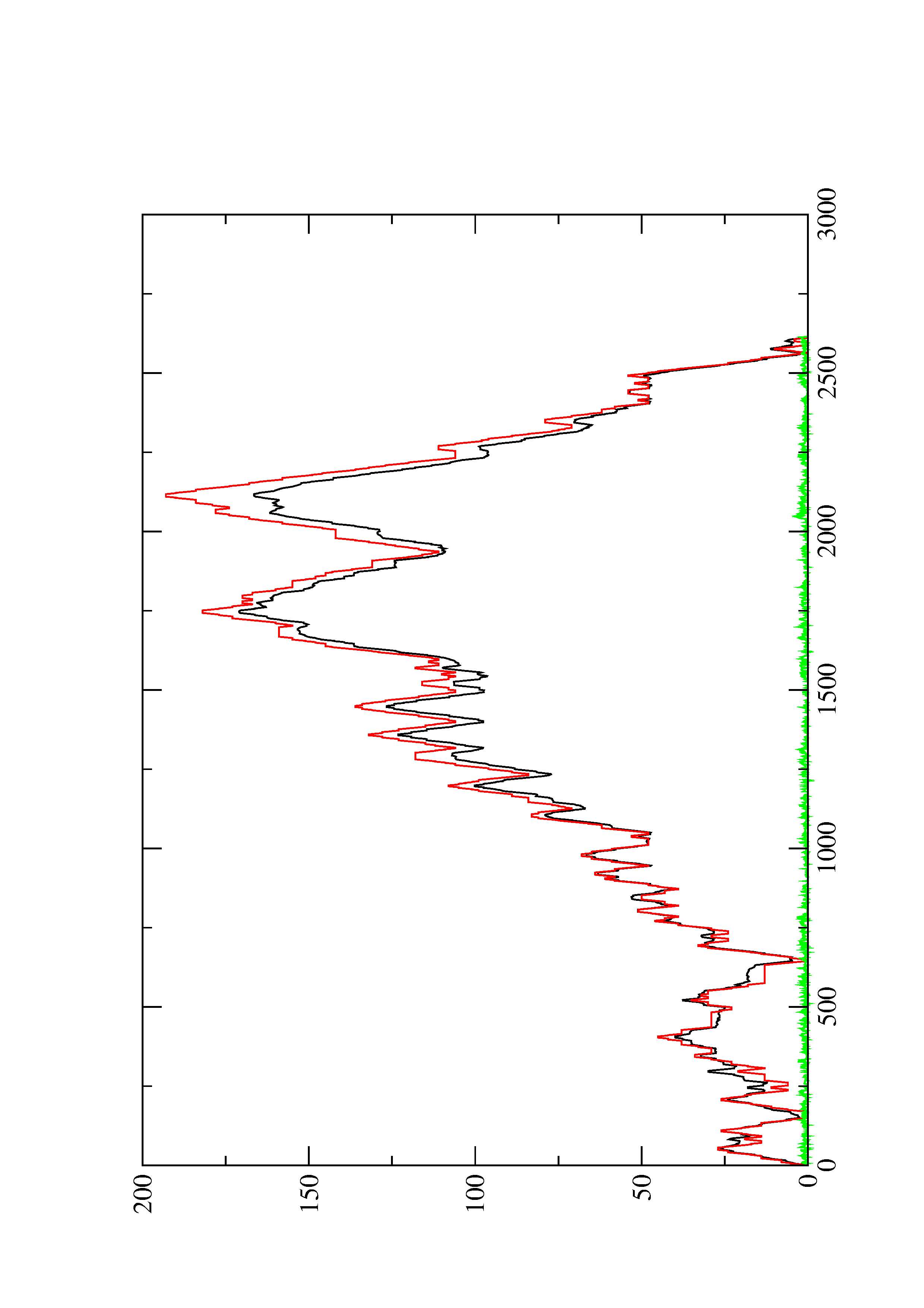
4. You may look at the dot plot containing the base pair probabilities [below]
III. Graphical output
1. A note on base-pairing probabilities:--
The structure below is colored by base-pairing probabilities. For unpaired regions the color denotes the probability of being unpaired. The reliability information of RNA secondary structure
plot in the form of color annotation, Positional entropy ranging from red (low entropy, well-defined) via green to blue and
violet (high entropy, ill-defined).
2. Here you find a mountain plot representation of the MFE structure, the thermodynamic ensemble of RNA structures, and the centroid structure. Additionally we present the positional entropy for each position. The resulting plot shows three curves, two mountain plots derived from the MFE structure (red) and the pairing probabilities (black) and a positional entropy curve (green). Well-defined regions are identified by low entropy.